
Christopher Petzold and Alyssa Redding-Johanson were key members of a team that demonstrated a technique called “targeted proteomics” to speed up and improve the identification and quantification of proteins within a sample. (Photo by Roy Kaltschmidt, Berkeley Lab Public Affairs)
Efforts to engineer new metabolic pathways into microbes for the inexpensive production of valuable chemical products, such as biofuels or therapeutic drugs, should get a significant boost in a new development from the U.S. Department of Energy (DOE)’s Joint BioEnergy Institute (JBEI). Researchers there have successfully demonstrated a technique they call “targeted proteomics” that speeds up and improves the ability to identify and quantify specific proteins within a cell or microorganism.
“Metabolic engineers and synthetic biologists can use our directed proteomic technique to get useful information about protein levels in their organisms, which in turn can be used to direct valuable followup experiments,” says Christopher Petzold, chemist and deputy director for proteomics at JBEI, who led this research. “We believe that targeted proteomics is a useful tool that fills a much needed gap in efforts to engineer new metabolic pathways for microbes.”
Petzold, who also holds an appointment with the Lawrence Berkeley National Laboratory (Berkeley Lab)’s Physical Biosciences Division, is the corresponding author on a paper describing this research that was published in the journal Metabolic Engineering. The paper is titled “Targeted proteomics for metabolic pathway optimization: Application to terpene production.” Coauthoring the paper with Petzold were Alyssa Redding-Johanson, Tanveer Batth, Rossana Chan, Rachel Krupa, Heather Szmidt, Paul Adams, Jay Keasling, Taek Soon Lee and Aindrila Mukhopadhyay.
Metabolic engineering is the practice of altering genes and chemical pathways within a cell or microorganism to increase production of a specific chemical substance. It is fast becoming one of the principal techniques of modern biotechnology for the microbial production of chemicals that are currently derived from non-renewable resources or from natural resources that are limited. Critical to the success of metabolic engineering efforts are techniques that enable researchers to assemble and optimize novel metabolic pathways in microbes. Given that such pathways often involve multiple different factors, performance- hampering problems, such as the abundance of proteins and messenger RNA, or the activity of enzymes, are not always evident simply by measuring the amount of final product obtained.
“Synthetic biologists and metabolic engineers will utilize a variety of analytical methods to identify the parts of a metabolic pathway that limit production,” Petzold says. “At the metabolite level, for example, monitoring all pathway intermediates helps identify bottlenecks where further engineering could improve the final product titer. However, pathways that divert intermediates to native processes at the cost of final product formation may disguise the actual location of a bottleneck.”
Researchers have tried developing assays to evaluate every intermediate in a given metabolic pathway but this has been a major challenge because intermediates often degrade rapidly or are isomers, and because there are few available standards for such evaluations.
“Engineers are often left to guess at the metabolite levels in parts of the pathway,” Petzold says.
Targeted proteomics is based on a variation of mass spectrometry called selected-reaction monitoring (SRM) that can be used to rapidly detect and quantify multiple target proteins within complex protein mixtures, such as those found in cells or microbes. When coupled to liquid chromatography (LC),SRM mass spectrometry analysis provides high selectivity and sensitivity through the elimination of background signal and noise even in the most complex mix of proteins. This is made possible by selecting only two points of mass for monitoring – a peptide mass and a specific fragment mass – rather than scanning the entire mass range.
“Carrying out targeted proteomics through SRM mass spectrometry analysis is most useful when quantification of multiple proteins in a single sample is desired,” Petzold says. “Working with protein-specific peptides, you can analyze 20 or more targeted proteins in an hour, and entire engineered metabolic pathways can be quantified in a single experiment, something that isn’t practical with conventional immunoblot analysis.”
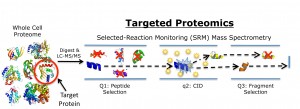
Schematic of targeted proteomics technique in which a peptide mass and a specific fragment mass are selected for SRM mass spectrometry analysis to identify and quantify a target protein. (Image from Christopher Petzold)
Petzold and his co-authors demonstrated the effectiveness of their targeted proteomics technique when they used it to measure protein levels in Escherichia coli that were engineered with yeast proteins to produce amorpha-4,11-diene, a member of the family of plant chemicals known as sesquiterpenes. Strains of E. coli containing a high flux mevalonate pathway have the potential to provide a vast range of high value sesquiterpenes and other isoprenoid-based chemical compounds, which today are typically obtained from petrochemical or plant sources.
Our analysis identified two mevalonate pathway proteins, mevalonate kinase and phosphomevalonate kinase, both from yeast, as potential bottlenecks,” Petzold says. “Codon-optimization of the genes encoding mevalonate kinase and phosphomevalonate kinase, and expression from a stronger promoter led to significantly improved levels of these two proteins and a more than three-fold improvement in the final amorpha-4,11-diene titer, greater than 500 milligrams per liter.”
Petzold and his JBEI colleagues are now in the process of implementing “scheduled-SRM,” a variation of the SRM mass spectrometry analysis that would allow them to easily detect and quantify 100 proteins in a single experiment that can be completed in less than two hours.
This research was supported by JBEI through the DOE Office of Science.
JBEI is one of three Bioenergy Research Centers funded by the U.S. Department of Energy to advance the development of the next generation of biofuels. It is a scientific partnership led by Berkeley Lab and including the Sandia National Laboratories, the University of California campuses of Berkeley and Davis, the Carnegie Institution for Science, and the Lawrence Livermore National Laboratory.
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 12 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
Additional information:
For more information about the Joint BioEnergy Institute, visit the Website at www.jbei.org