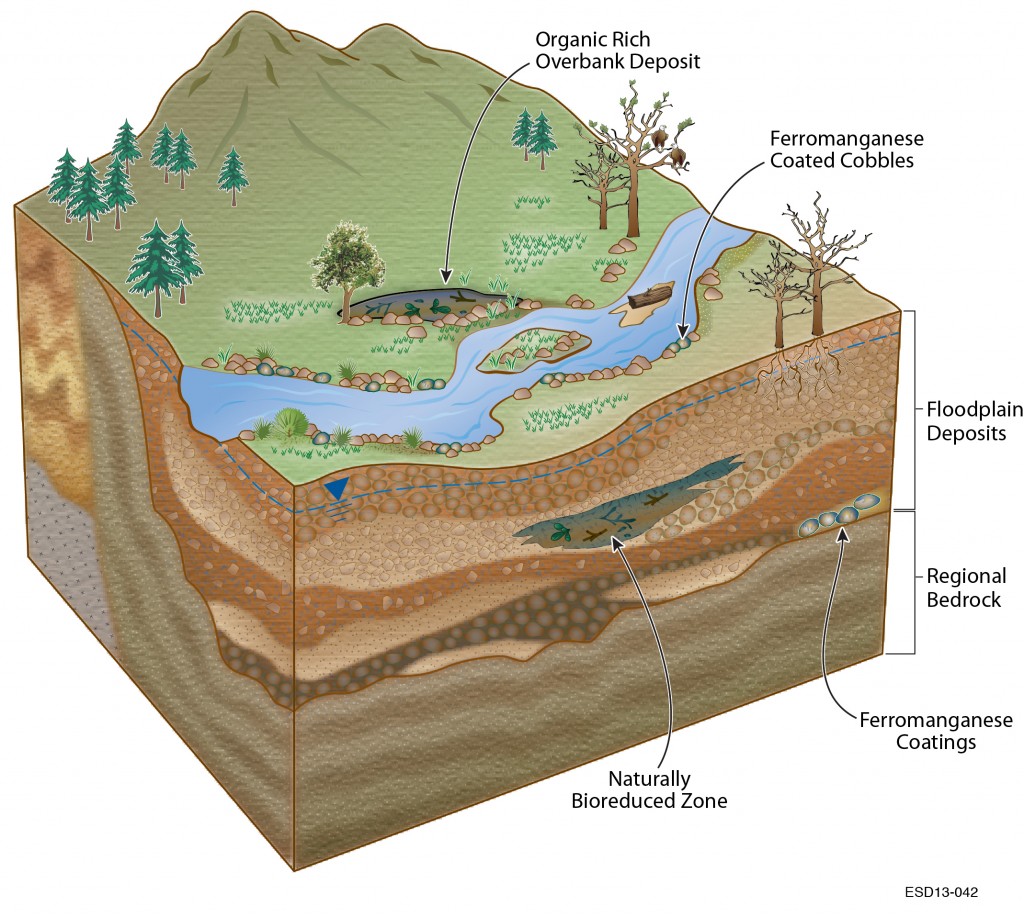
The key to a better understanding of the carbon cycle, the flow of contaminants, even the sustainable growth of biofuel crops, starts with the ground beneath your feet. More specifically, it starts with the genomes of the microbes that live in the water and sediment beneath your feet.
But that’s where things get murky. It turn out that many of the resources we depend on—clean water, food, and energy—in turn rely on subsurface microbial functions that are poorly understood or have yet to be discovered. Scientists estimate that more than half the branches on the bacterial tree of life are unknown. What these microbes do in the subsurface, and how they take part in important biogeochemical processes, is a mystery.
That could change, thanks to a new Berkeley Lab-led project that will develop a predictive understanding of how the genomic functions of subsurface microbiomes affect watershed-scale biogeochemical processes.
The project is called Sustainable Systems Scientific Focus Area 2.0, and involves 50 scientists from Berkeley Lab and other institutions including UC Berkeley, Pacific Northwest National Lab, and Oak Ridge National Lab. It’s led by geophysicist Susan Hubbard, who is also the director of Berkeley Lab’s Earth Sciences Division.
“This genome-through-watershed scale project will be the first to develop approaches for interrogating and simulating how global change affects microbially induced biogeochemical cycling in the subsurface, as is relevant to carbon cycling, contaminant mobility, and agricultural sustainability,” says Hubbard.
The Sustainable Systems Scientific Focus Area 2.0 is supported by the Subsurface Biogeochemistry Program within the Department of Energy’s Office of Biological and Environmental Research.
Scientists will work to answer several big unknowns: How will climate and land-use induced changes in hydrology and vegetation affect subsurface carbon inputs, flowpaths and subsurface metabolic potential? How will these processes evolve over time? And what effect will these interactions have on how watershed-scale biogeochemical processes play out over time?
Developing a predictive understanding of these processes and their interactions will provide a much needed critical underpinning for a wide range of environmental problems.

Berkeley Lab’s Tetsu Tokunaga and Yongman Kim install vertically resolved vadose zone samplers at the Rifle Research Site, with silt-rich surface materials and alluvium on display.
The project will require development of new process knowledge, terabase-scale sequencing from DOE’s Joint Genome Institute, capabilities to manage and mine diverse environmental datasets, genome-enabled watershed simulators, extensive field research at key locations such as the Rifle Research Site in Colorado, and environmental and biological systems expertise.
To get a sense of the kind of scientific dividends this wide-ranging collaboration will yield, consider recent findings from the lab of SFA 2.0 member Jill Banfield, which were developed as the project was spinning up. Banfield, a geomicrobiologist, is a Senior Faculty Scientist in Berkeley Lab’s Earth Sciences Division, and a UC Berkeley professor in the Departments of Earth and Planetary Sciences and Environmental Science, Policy and Management.
Her team has been conducting large-scale sequencing of DNA extracted from the aquifer at the Rifle site, reconstructing the genomes of the organisms present, and finding an incredibly diverse mix of unknown and previously unstudied organisms. Their findings, which have been documented through a string of high-impact publications, include:
- New insights into RuBisCO, an enzyme involved in carbon fixation. It’s one of the most abundant enzymes on the planet and one of the best studied. But in a study published in the journal Science, UC Berkeley’s Kelly Wrighton et al found that previously unstudied bacterial phyla use a novel form of RuBisCO, likely for carbon fixation.
- Genomes from unknown lineages of organisms involved in carbon and nitrogen cycling. As published in the journal Microbiome, UC Berkeley’s Laura Hug et al used large-scale sequencing techniques to explore the diversity and variety of a bacterial phylum called Chloroflexi, and to study its role in carbon cycling.
- The extraordinary diversity of an aquifer’s microbiome. In a study published in Nature Communications, UC Berkeley’s Cindy Castelle et al reported that terabase sequencing of aquifer samples uncovered an incredible diversity of life, including previously unknown phyla, and the complete genome of a microbe from one of these new phyla.
“This is just the tip of the iceberg. We are at the very beginning stages of exploring the diversity of subsurface life,” says Banfield. “This SFA will advance our understanding of subsurface ecogenomics and eventually lead to predicting how the subsurface microbiome interacts with and influences environmental systems and biogeochemical cycling. Development of these new capabilities will be transformational for environmental science.”
###
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.