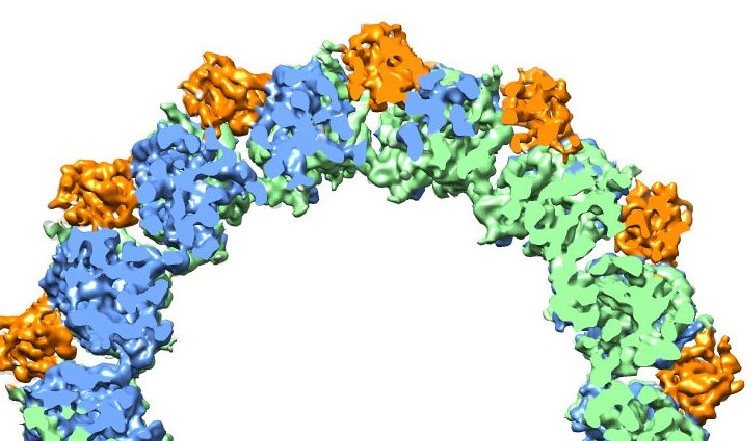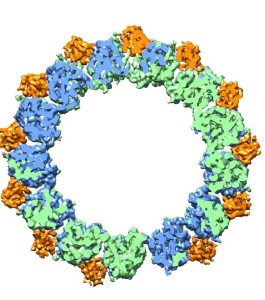
Microtubules are hollow cylinders with walls made up of tubulin proteins – alpha (green) and beta (blue) – plus EB proteins (orange) that can either stabilize or destabilize the structure of the tubulin proteins.
Microtubules, hollow fibers of tubulin protein only a few nanometers in diameter, form the cytoskeletons of living cells and play a crucial role in cell division (mitosis) through their ability to undergo rapid growth and shrinkage, a property called “dynamic instability.” Through a combination of high-resolution cryo-electron microscopy (cryo-EM) and a unique methodology for image analysis, a team of researchers with Berkeley Lab and the University of California (UC) Berkeley has produced an atomic view of microtubules that enabled them to identify the crucial role played by a family of end-binding (EB) proteins in regulating microtubule dynamic instability.
During mitosis, microtubules disassemble and reform into spindles that are used by the dividing cell to move chromosomes. For chromosome migration to occur, the microtubules attached to them must disassemble, carrying the chromosomes in the process. The dynamic instability that makes it possible for microtubules to transition from a rigid polymerized or “assembled” nucleotide state to a flexible depolymerized or “disassembled” nucleotide state is driven by guanosine triphosphate (GTP) hydrolysis in the microtubule lattice.
“Our study shows how EB proteins can either facilitate microtubule assembly by binding to sub-units of the microtubule, essentially holding them together, or else cause a microtubule to disassemble by promoting GTP hydrolysis that destabilizes the microtubule lattice,” says Eva Nogales, a biophysicist with Berkeley Lab’s Life Sciences Division who led this research.

Gregory Alushin and Eva Nogales studying images of microtubule structures. (Photo by Roy Kaltschmidt)
Nogales, who is also a professor of biophysics and structural biology at UC Berkeley and investigator with the Howard Hughes Medical Institute, is a leading authority on the structure and dynamics of microtubules. In this latest study, she and her group used cryo-EM, in which protein samples are flash-frozen at liquid nitrogen temperatures to preserve their natural structure, to determine microtubule structures in different nucleotide states with and without EB3. With cryo-EM and their image analysis methodology, they achieved a resolution of 3.5 Angstroms, a record for microtubules. For perspective, the diameter of a hydrogen atom is about 1.0 Angstroms.
“We can now study the atomic details of microtubule polymerization and depolymerization to develop a complete description of microtubule dynamics,” Nogales says.
Beyond their importance to our understanding of basic cell biology, microtubules are a major target for anticancer drugs, such as Taxol, which can prevent the transition from growing to shrinking nucleotide states or vice versa.

Rui Zhang is the lead author of a Cell paper describing the record 3.5 Angstroms resolution imaging of microtubules.
“A better understanding of how microtubule dynamic instability is regulated could open new opportunities for improving the potency and selectivity of existing anti-cancer drugs, as well as facilitate the development of novel agents,” Nogales says.
Nogales is the corresponding author of a paper describing this research in the journal Cell. The paper is entitled “Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins.” Co-authors are Rui Zhang, Gregory Alushin and Alan Brown.
This work was funded by a grant from NIH’s National Institute of General Medical Sciences.
Additional Information
For more about the research of Eva Nogales go here