-Written by Glenn Roberts
Scientists have for the first time viewed how bacterial proteins self-assemble into thin sheets and begin to form the walls of the outer shell for nano-sized polyhedral compartments that function as specialized factories.
The research, led by researchers at the U.S. Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) and Michigan State University in collaboration with the University of Liverpool, provides new clues for scientists seeking to use these 3-D structures as “nanoreactors” to selectively suck in toxins or churn out desired products.
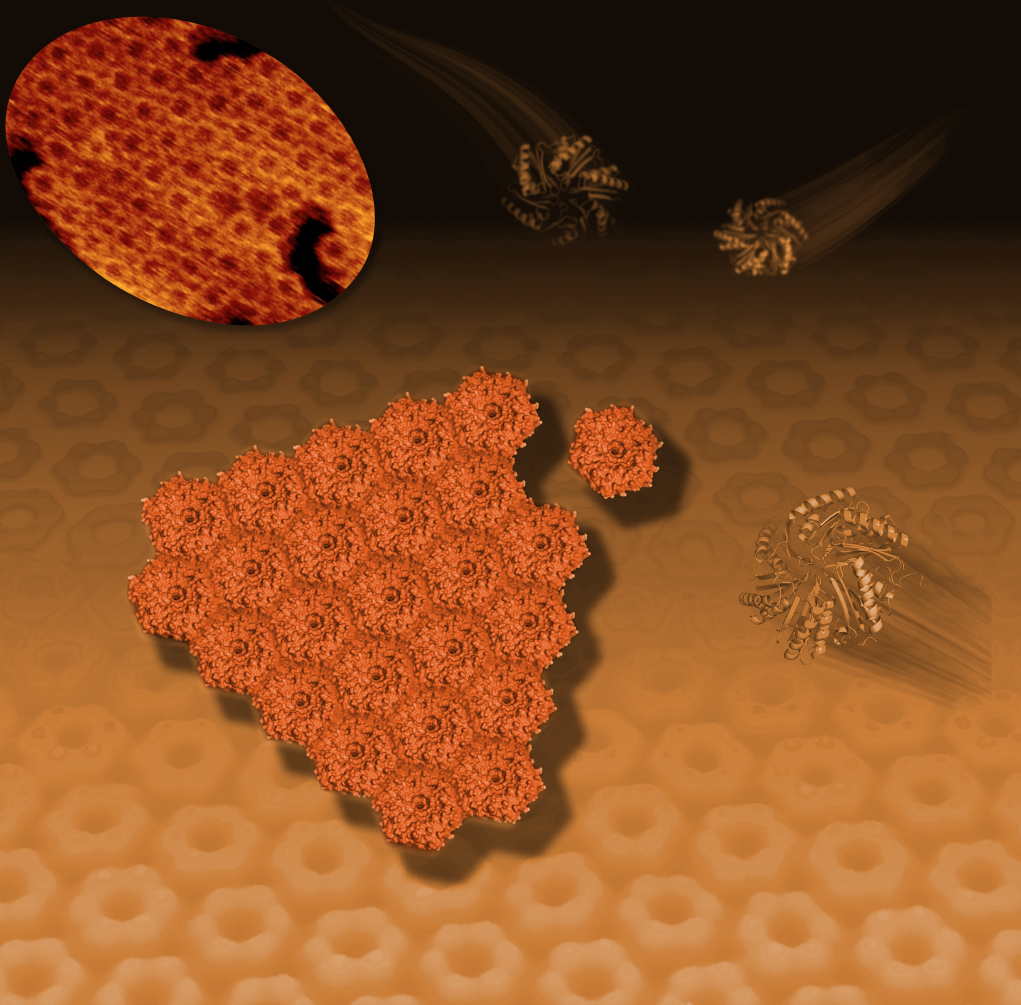
This illustration shows how hexagonal bacterial proteins (shown as ribbon-like structures at right and upper right) self-assemble into a honeycomb-like tiled pattern (center and background). This tiling activity, seen with an atomic-resolution microscope (upper left), represents the early formation of polyhedral, soccer-ball-like structures known as bacterial microcompartments or BCMs that serve as tiny factories for a range of specialized activities.
The new insight may aid scientists who seek to tap this natural origami by designing novel compartments or using them as scaffolding for new types of nanoscale architectures, such as drug-delivery systems.
“We have a new clue in understanding nature’s inner-cell architecture,” said Cheryl Kerfeld, a Berkeley Lab structural biologist who is co-corresponding author on the study. Her research group at Berkeley Lab specializes in the structure and inner workings of these tiny compartments, known as bacterial microcompartments or BMCs. Kerfeld holds joint appointments with Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging (MBIB) Division and Michigan State University.
“We usually only get to see these structures after they form, but in this case we’re watching them assemble and answering some questions about how they form,” Kerfeld said. “This is the first time anyone has visualized the self-assembly of the facets, or sides, of the microcompartments. It’s like seeing walls, made up of hexagonally shaped tiles, being built by unseen hands.”
The study was published online Nov. 30 in Nano Letters.
Several models had been proposed for how these compartments are built from scratch inside bacteria by proteins, and there were many open questions about the construction process.
Researchers combined X-ray studies of the 3-D structure of a protein that resembles a hexagon with imaging by an atomic-force microscope to reveal how the hexagons arrange in a honeycomb pattern in the microcompartment’s walls.
Markus Sutter, a Berkeley Lab scientist who is the study’s lead author, determined the 3-D structure of the basic building block protein at the Advanced Light Source at Berkeley Lab using crystallized samples. Patterns produced when X-rays struck the protein crystals provided key details about the protein’s shape, at the scale of individual atoms. “That gave us some exact dimensions,” Sutter said, which helped to interpret the microscope images. “It also showed us that hexagons had distinct sidedness: One side is concave, the other side is convex.”
Markus Sutter, a Berkeley Lab scientist, determined the 3-D atomic structure of a bacterial protein that self-assembles into honeycomb-patterned sheets using X-rays at beamline 5.0.1 (pictured here) at Berkeley Lab’s Advanced Light Source. (Photo by Roy Kaltschmidt)
Liverpool’s atomic-force microscope, BioAFM, showed that individual hexagon-shaped protein pieces naturally join to form ever-larger protein sheets in a liquid solution. The hexagons only assembled with each other if they had the same orientation—convex with convex or concave with concave.
“Somehow they selectively make sure they end up facing the same way,” Kerfeld added.
The study also found that individual hexagon-shaped pieces of the protein sheet can dislodge and move from one protein sheet to another. Such dynamics may allow fully formed compartments to repair individual sides.
The protein sheets studied were not viewed inside living bacteria, though the conditions of the microscope experiment were designed to mimic those of the natural bacterial environment. “We think this is what goes on when these compartments assemble inside the microbe,” Kerfeld said.
Some studies have proposed that the protein shell of microcompartments might be several layers thick. However, this study suggests that the shell facets are composed of a single protein layer. Sutter said this makes sense: The compartments are known to selectively allow some chemical exchanges between their contents and their outside environment, and a thicker shell could complicate these exchanges.
The exact mechanism for this chemical exchange is not yet well-understood. This and other mysteries of the microcompartments can hopefully be resolved with follow-up studies that seek to chronicle the complete assembly process, the researchers said.
Fully-formed 3-D microcompartments have a soccer-ball-like geometry that incorporates pentagon-shaped protein structures known as pentamers, for example, that were not included in the latest study.
“The holy grail is to see the structure and dynamics of an intact shell, composed of several different types of hexagonal proteins and with the pentagons that cap its corners,” Kerfeld said.
It’s possible that simply adding these pentamers to the protein sheets from the latest experiment could stimulate the growth of a complete 3-D structure, but Kerfeld added, “I wouldn’t be surprised if there’s more to the story.”
Once more is learned about the microcompartments, it’s conceivable they could be used to concentrate the production of beneficial enzymes, organize them to produce an ordered sequence of chemical reactions, or to remove particular toxins from the surrounding environment, she said.
The X-ray studies were performed at Berkeley Lab’s Advanced Light Source, a DOE Office of Science User Facility, and the microscope work was performed by Kerfeld’s collaborator, Luning Liu at the University of Liverpool.
Matthew Faulkner, Clément Aussignargues, Bradley C. Paasch and Steve Barrett also participated in the study.
This research was supported by the National Institutes of Health.
Additional Information
For more about the research of Cheryl Kerfeld go here.
# # #
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit the Office of Science website at science.energy.gov/.