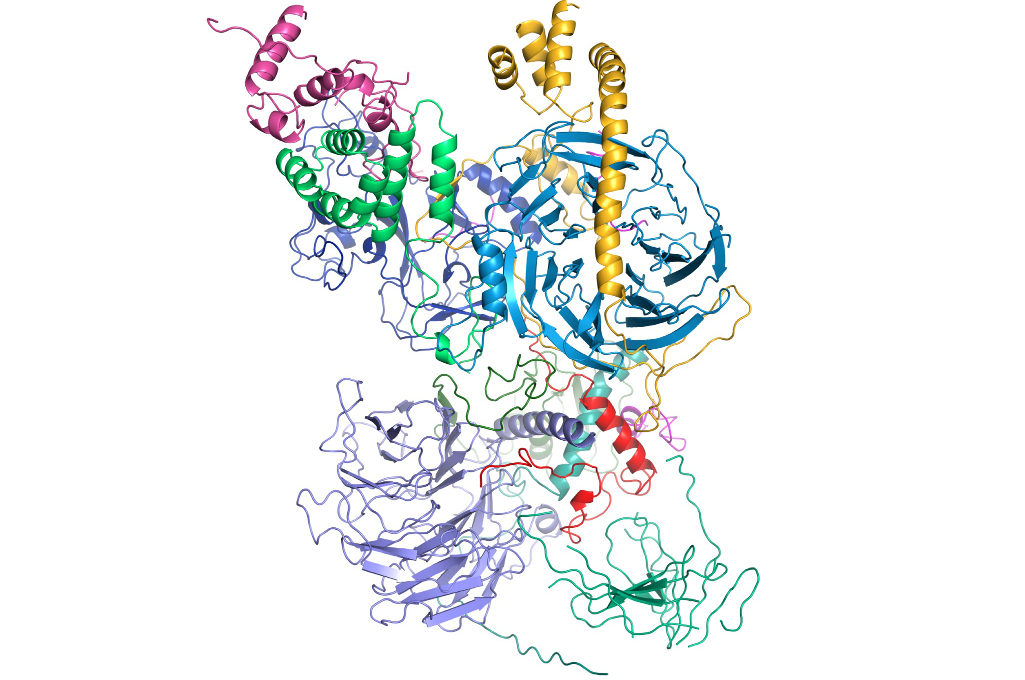
Structure of the human Polycomb Repressive Complex 2 (PRC2) bound to cofactors obtained by cryo-electron microscopy. Both cofactors mimic the histone protein tail to stabilize and stimulate the enzymatic activity of PRC2. (Credit: Vignesh Kasinath)
All the trillions of cells in our body share the same genetic information and are derived from a single, fertilized egg. When this initial cell multiplies during fetal development, its daughter cells become more and more specialized. This process, called cell differentiation, gives rise to all the various cell types, such as nerve, muscle, or blood cells, which are diverse in shape and function and make up tissues and organs. How can the same genetic blueprint lead to such diversity? The answer lies in the way that genes are switched on or off during the course of development.
Scientists at Lawrence Berkeley National Laboratory (Berkeley Lab) have been studying the molecules that act at the genetic level to give rise to different types of cells. Some of these molecules are a complex of proteins called the Polycomb Repressive Complex 2 (PRC2) that is involved in “silencing” genes so that they are not “read” by the cellular machinery that decodes genetic information, effectively keeping the genetic information in the “off” state.
In two new studies, a team of researchers led by Eva Nogales, senior faculty scientist in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division, has gained insight into the structure of PRC2 and the ways in which it is regulated to affect gene silencing. Their work was reported on January 18 in the journal Science and on January 29 in Nature Structural and Molecular Biology by Eva Nogales and postdoctoral researchers Vignesh Kasinath and Simon Poepsel.
Both publications provide a structural framework to understand PRC2 function, and in the case of the latter, the structures are the first to illustrate how a molecule of this type engages with its substrate. The structural descriptions of human PRC2 with its natural partners in the cell lend important insight into the mechanism by which the PRC2 complex regulates gene expression. This information could provide new possibilities for the development of therapies for cancer.
PRC2 is a gene regulator that is vital for normal development. Genomic DNA is packaged into nucleosomes, which are formed by histone proteins that have DNA wrapped around them. Histone proteins have long polypeptide tails that can be modified by the addition and removal of small chemical groups. These modifications influence the interaction of nucleosomes with each other and other protein complexes in the nucleus. The function of PRC2 in the cell is to make a particular chemical change in one of the histones. The genes in the regions of the genome that have been modified by PRC2 are switched off, or become silenced.
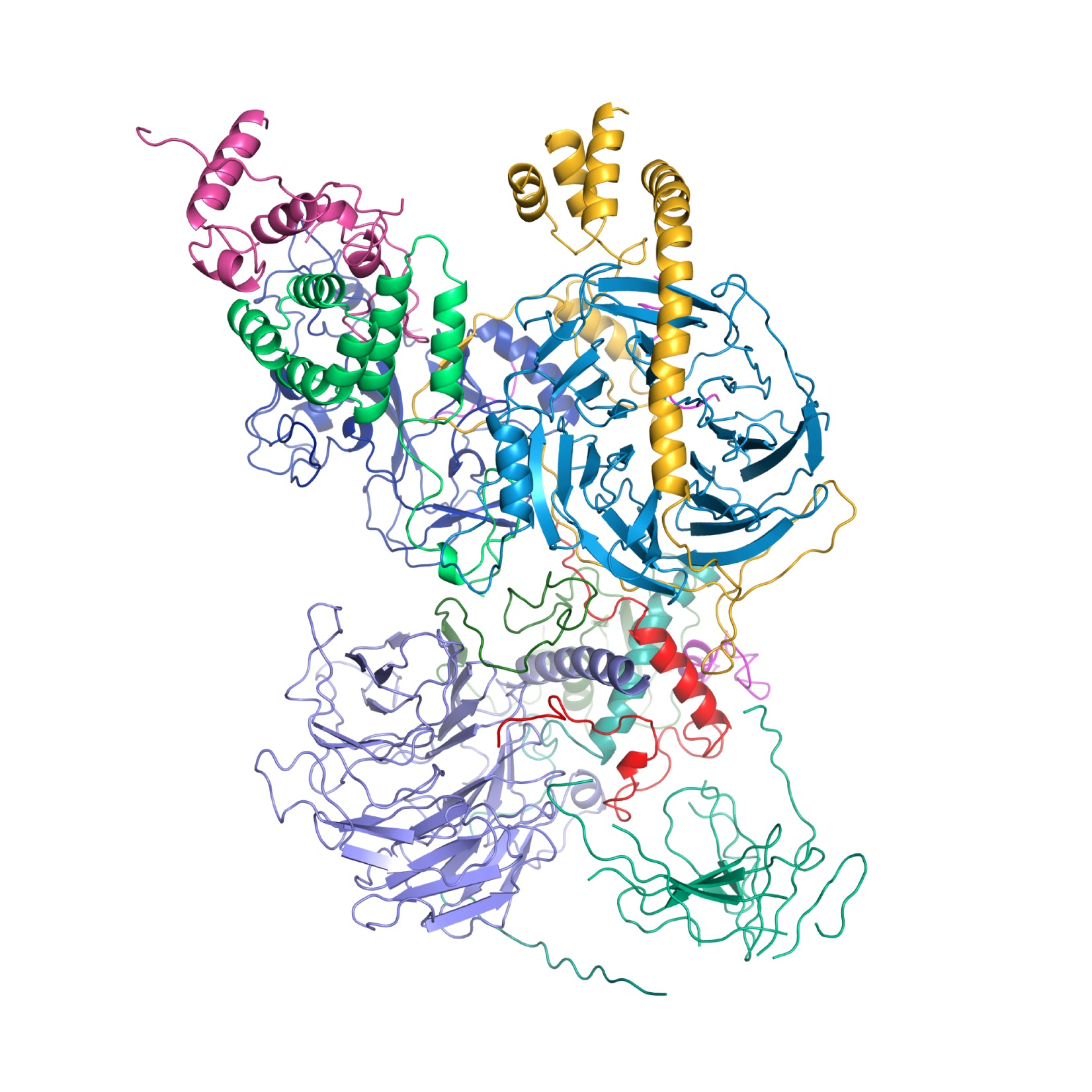
This montage of the full PRC2 with two nucleosomes is based on the superposition of the cryo-EM maps of PRC2 with and without the nucleosomes to show the consistency of the observed nucleosome binding configuration with the full PRC2 structure. (Credit: Simon Poepsel)
“Not surprisingly, elaborate mechanisms have evolved to ensure that PRC2 marks the correct regions for silencing at the right time,” said Nogales, who is also a Howard Hughes Medical Investigator and professor of Biochemistry, Biophysics and Structural Biology at the University of California, Berkeley. Failure of this regulation not only impairs the process of development, but also contributes to the reversal of cell differentiation and the uncontrolled cell growth that are the hallmarks of cancer. “Therefore,” Nogales continued, “gaining insight into how PRC2 function is adjusted both in space and time is crucial to understanding cell development.”
Nogales and her team use structural biology to elucidate how biomolecules, particularly proteins and nucleic acids (DNA, RNA), are organized and combine to form functional biological assemblies. Obtaining detailed insights into their three-dimensional shape will not only help to understand how they function but also how this function is regulated in the cell. These two studies rely on cryo-electron microscopy for imaging the biomolecules, a technique that can see large biomolecules on a very small scale and in multiple conformations. Kasinath and Poepsel, have now solved the structure of PRC2, which provides a framework to understand how this complex is regulated to modify histone proteins.
The first study, published January 18 in Science by Kasinath, Poepsel, Nogales, and coworkers, visualized the architecture of the complete PRC2 in atomic detail. First author Vignesh Kasinath said, “It took three years of work to obtain this high-resolution structure of all the parts, or subunits, that make up a functional PRC2, as well as visualize how additional protein subunits, called cofactors, may help regulate its activity. Remarkably, both cofactors mimic the histone protein tail in their binding to PRC2 suggesting that cofactors and histone tails together work hand-in-hand to regulate PRC2 function. This structural work holds great promise for new drug development to fight PRC2 dysfunction in cancer.”

From left, Simon Poepsel, Eva Noagles, and Vignesh Kasinath were part of a team of scientists that led the research. (Credit: Basil Greber)
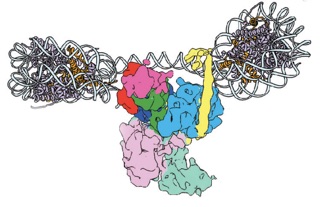
This work is complemented by a second study that presents snapshots of PRC2 binding to the histone proteins that it modifies as a signal for gene silencing. The structures, which have been published in Nature Structural and Molecular Biology on January 29 by Poepsel, Kasinath and Nogales this week, illustrate beautifully the action of this sophisticated complex. “PRC2 can simultaneously engage two nucleosomes,” said Poepsel, first author of this study. “Our cryo-EM images help us understand how the complex can recognize the presence of a histone modification in one nucleosome and place the same tag onto a neighboring nucleosome.” This cascade of activity enables PRC2 to spread this modification over the entire neighboring gene loci, thereby marking it for silencing. Nogales added, “The visualization of such interactions is notoriously hard. We have made an important step forward in our general understanding of how gene regulators can bind to and recognize nucleosomes.”
PRC2 is essential to gene regulation and expression in all multicellular organisms. The findings from both studies open up tremendous possibilities for combatting cancer while simultaneously expanding our knowledge of gene regulation at a molecular level. “Because PRC2 is deregulated in cancers, it makes a good target for potential therapeutics,” said Nogales. The fundamental understanding of PRC2 arising from these studies will have broad implications in both plant and animal biology.
This work was funded by the Howard Hughes Medical Institute and Eli Lilly. This research used cryo-electron microscopy (cryo-EM) and made use of the unique resources of the Bay Area Cryo-EM Facility. Image analysis relied on heavy computational work that was carried out at Berkeley Lab’s National Energy Research Scientific Computing Center (NERSC), a DOE Office of Science User Facility. Vignesh Kasinath was supported by a postdoctoral fellowship from Helen Hay Whitney and Simon Poepsel was supported by the Alexander von Humboldt foundation (Germany) as a Feodor-Lynen postdoctoral fellow.
###
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel Prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.