Contact: Linda Vu, (510) 495-2402, <[email protected]>
“We live in a microbial world,” says Nikos Kyrpides of Berkeley Lab’s Genomics Division. “There are millions of organisms in one drop of water and even more in soil. Life on our planet cannot be sustained without the microbes.”
However, only a tiny fraction of microbes live as independent species, and even fewer of these can be cultured in the laboratory. The vast majority of bacteria and other microorganisms exist only in the wild, and in complex communities. The collective genome of such a microbial community, its total DNA, is called its metagenome.
To make sense of these metagenomes, scientists rely on analytic tools like the Integrated Microbial Genomics with Microbiome Samples (IMG/M) – which is a cumulative database that includes individual gene sequences, partial and whole genomes from individual organisms, and other DNA and RNA sequences recovered from wild communities.
IMG/M was developed through a close collaboration of software engineers, computer scientists, and biologists from the Genome Biology and Microbial Ecology programs of the U.S. Department of Energy’s Joint Genome Institute (JGI), as well as the Biological Data Management and Technology Center (BDMTC) in Berkeley Lab’s Computational Research Division. IMG/M has played a central role in helping scientists understand metagenomes in a variety of natural environments since its initial release in 2006.
Now a new grant from the National Institutes of Health (NIH) will expand the system’s capabilities to include metagenomic data from humans, giving scientists valuable insights into how microbial communities affect human health.
“The success of metagenomics will not only help us better understand human health, but may also help us address a variety of environmental challenges,” says Kyrpides, who heads JGI’s Genome Biology Program.
IMG/M, created under the auspices of DOE’s Office of Biological and Environmental Research, started at Berkeley Lab as a Laboratory Directed Research and Development program in 2005. The system was released in 2006, with Victor Markowitz, head of BDMTC, as IMG/M’s technical lead.
Supporting the Human Microbiome Project
“When the average person hears the word ‘microbe,’ they think of a disease or a disaster,” says Kyrpides. “However, the vast majority of microbes are our friends. In fact, entire microbial communities work in harmony with us to carry out essential functions, such as digestion in the human gut.”
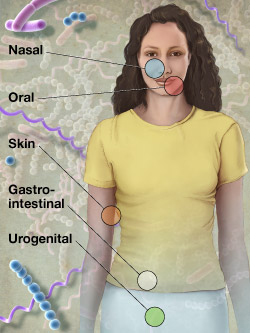
Within the body of a healthy adult, microbial cells are estimated to outnumber human cells by ten to one. These tiny organisms cover every surface and cavity of the human body, forming complex communities that help digest food, break down toxins, and fight off diseases.
“When these communities are disturbed, people may get sick or catch infections,” says Kyrpides. “Microbes have won every major battle on our planet – except that of making a good impression.”
To understand how microbes affect human health and how they cause various diseases, researchers involved in NIH’s Human Microbiome Project will collect metagenome samples from individuals with a variety of health conditions and from different parts of the human body. They will then use IMG/M to analyze the metagenome datasets generated from these samples.
The field of metagenomics is relatively new, Kyrpides says. Until a few years ago scientists studied individual microbes by growing them in laboratories, extracting their DNA, and then examining the sequence of their genes in order to understand the organism’s genetic makeup. While this approach was somewhat successful, he notes that it had substantial limitations, because most microbes cannot be grown in laboratories.
When scientists extract DNA from an entire microbiome sample, containing potentially hundreds of different microbial species, at first they don’t necessarily know which individual organism the genes come from or the function these genes carry out in the context of the community. This is the challenge of metagenomics and also its power: piecemeal, little by little, the various players in microbial communities become known, the abilities of their dominant members can be identified, and the genes that confer these abilities are specified and added to the database, even if complete genomes of most of the species are never finished.
“The IMG/M system is an invaluable tool in the quest of finding how communities function,” says Kyrpides. “The system allows us to analyze metagenomic datasets in the rich context of all available individual microbial genomes, and provides scientists with tools to compare and identify the functional capabilities of microbial communities.”
This past year, researchers used IMG/M to learn how microbes in Seattle’s Lake Washington enable the oxidation of methane, methanol, and methylated amines, compounds contributing to the greenhouse effect and the global carbon cycle.
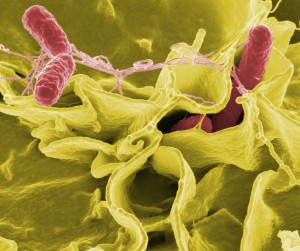
Color-enhanced scanning electron micrograph showing Salmonella typhimurium (red) invading cultured human cells. (Credit: Rocky Mountain Laboratories,NIAID,NIH)
It was the system’s track record in analyzing metagenomes from these types of natural environments that inspired scientists working on the Human Microbiome Project to include IMG/M in their NIH proposal to create a Data Analysis Coordination Center (DACC). This center will act as a central repository for all the human metagenome data collected by the project.
Greengenes, a website used by biologists to detect and classify microorganisms based on DNA samples, will also be part of the DACC. The greengenes system was developed by a team from Berkeley Lab’s Earth Sciences Division led by Gary Andersen.
Says Markowitz, “We are thrilled that two Berkeley Lab resources will support an initiative of such magnitude. We are looking forward to enhancing their capabilities through a joint effort of scientists from three different divisions.”
The principal investigator on the NIH grant is Owen White of the Institute for Genome Sciences at the University of Maryland’s School of Medicine in Baltimore. In addition to Berkeley Lab’s Kyrpides, Markowitz, and Andersen, investigators include Robin Knight of the Department of Chemistry and Biochemistry at the University of Colorado in Boulder.
Additional information
More about Integrated Microbial Genomics with Microbiome Samples (IMG/M) is at http://www.lbl.gov/CS/Archive/news053006b.html
More about the Biological Data Management and Technology Center at Berkeley Lab’s Computational Research Division is at http://crd.lbl.gov/html/BDMTC/index.html
More about the Genome Biology Program at DOE’s Joint Genome Institute is at http://www.jgi.doe.gov/
For more information on the Human Microbiome Project’s Data Analysis Coordination Center (DACC), please visit http://nihroadmap.nih.gov/hmp
For more information on the greengenes project, visit http://greengenes.lbl.gov/cgi-bin/nph-index.cgi
For more about Nikos Kyrpides visit http://www.jgi.doe.gov/research/kyrpides.html
For more about Victor M. Markowitz visit http://crd.lbl.gov/html/BDMTC/markowitz.html
For more about Gary Andersen visit http://www-esd.lbl.gov/ECO/MME/staff_andersen.htm