If you keep up with biology, you’ve probably seen those colorful images in which the atom-by-atom structure of a protein is portrayed by a tangle of ribbons. For the past couple of decades, scientists have been hard at work deriving these structures, which provide clues as to what proteins do.
It’s critical research. Proteins are workhorse compounds that sustain all living things. The next drug breakthrough, or insight into how life works at the molecular scale, could originate from one of these images.
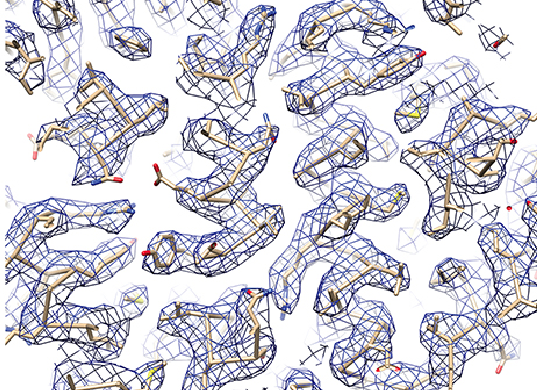
UC San Francisco scientists used cryo-EM to determine the 3.3-Å-resolution structure of a microbe’s 20S proteasome, which degrades unneeded or damaged proteins. This image depicts a portion of a cryo-EM density map showing clear side-chain densities. (Source: Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM, Li, et al, Nature Methods. (2013) 10:584-590.)
Most protein structures are obtained via x-ray crystallography. Scientists also use nuclear magnetic resonance imaging to solve structures. And coming in at a distant third is a technique called single-particle cryoelectron microscopy (cryo-EM), in which a macromolecule is immobilized in a thin film of vitrified buffer and inserted into the vacuum of an electron microscope.
Cryo-EM’s back-of-the-pack status is changing, however, thanks in part to technology developed at Berkeley Lab.
The shift is due to advances in the camera, or detector. Traditional high-performance electron microscopy cameras record the image indirectly. Electrons hit a scintillator, and the resulting light is sent to a CCD chip, where it is converted to an electronic image and read-out.
But a new generation of cameras offers a more direct approach. With these cameras, electrons hit the silicon pixels directly, and the resulting events are collected by microfabricated electronics that render an image.
One such direct-conversion electron detector owes its origins to a camera developed for one of the world’s most powerful electron microscopes, TEAM, which is located at Berkeley Lab’s National Center for Electron Microscopy. The camera was engineered by Berkeley Lab’s Peter Denes and colleagues, and began its service when TEAM started operating in 2009.
A version of the Berkeley Lab camera has since been commercialized by the Pleasanton, California-based Gatan, Inc. Its 16 million pixels capture events at 400 frames per second. Several are now in use at UC San Francisco, Caltech, and in the UC Berkeley lab of Eva Nogales, who is also a scientist in Berkeley Lab’s Life Sciences Division.
This camera, along with two other commercially available direct-conversion electron cameras, are making cryo-EM a real player when it comes to mapping protein structures.
For example, scientists from the United Kingdom used the method to derive a 4.5-angstrom resolution image of the yeast ribosome, and did so using only two percent of the data than was necessary for earlier efforts that produced lower-resolution results. Previously, scientists had to merge data from as many as a couple of million single particles in order to improve the signal-to-noise ratio, and even then they were not able see ribosomal structures such as side chains. Now, only 35,000 particles are needed.
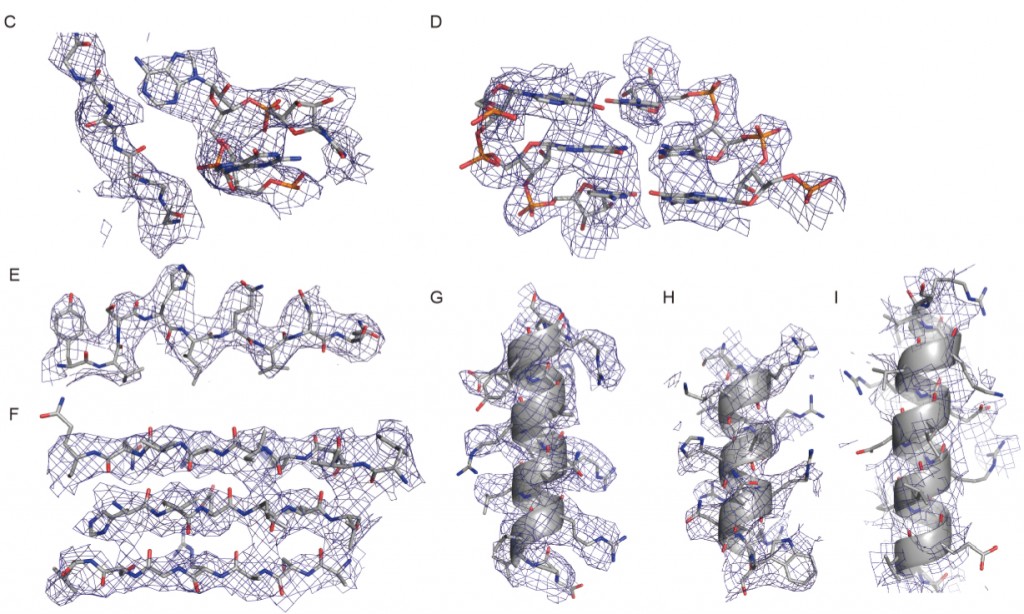
Scientists from the United Kingdom’s Medical Research Council Laboratory of Molecular Biology used cryo-EM to solve ribosome structures of the yeast ribosome to near-atomic resolution. In these images: Densities for the 60S subunit a protein loop interacting with a flipped-out RNA base (C), a short stretch of an RNA helix (D), a β-strand (E), a β-sheet (F), and an α-helix (G). (H–I) Densities for the 40S subunit showing a well-resolved α-helix. Figure I is not well resolved. (Credit: Sjors Scheres, Medical Research Council Laboratory of Molecular Biology)
“That’s a huge and sudden improvement,” says Robert Glaeser, a scientist in Berkeley Lab’s Life Sciences Division and a pioneer in using cryo-EM to determine the structure of proteins. Glaeser outlined the impacts of direct-conversion cameras in a recent News and Views article for Nature Methods.
In addition, as Glaeser outlines in the Nature Methods article, the new, direct-conversion camera makes it possible to record a ‘stroboscopic movie’ while exposing a protein to electrons. Such movies are important because the cryo-EM specimen constantly moves while an image is being recorded, probably because of stresses caused by radiation damage.
Improvements like these put many important proteins within reach for the first time. Recently, scientists from UC San Francisco used cryo-EM to solve the structure of a much smaller protein complex than previously possible with the technique: the 20S proteasome. And Nogales is using the technique to derive high-resolution structures of a microtubule, a protein that helps to maintain the structure of a cell among other functions.
There are several advantages to using cryo-EM for deriving protein structures. Unlike x-ray crystallography, a protein doesn’t have to be crystallized in order to map it, which is a time consuming process. And a protein’s structure can be determined in its biochemically native solution.
So, the next time you see an image of a near atomic-scale structure of a protein, be sure to read the caption. Don’t be surprised if it’s derived from single-particle cryo-EM.
###
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.