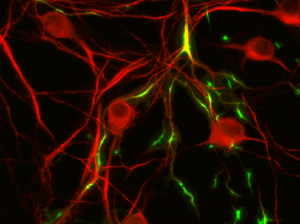
TREK1 potassium ion channels, fluorescing green in these cultured neurons, have been linked to the regulation of emotions. (Image from Isacoff group)
Using a unique and relatively simple cell-based fluorescent assay they developed, scientists with the U.S. Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab) and the University of California (UC), Berkeley have identified a means by which fluoxetine, the active ingredient in Prozac, suppresses the activity of the TREK1 potassium channel. TREK1 activity has been implicated in mood regulation and could be an important target for fluoxetine and other antidepressant drugs.
“Whereas the inhibiting of serotonin re-uptake remains fluoxetine’s primary antidepression mechanism, many pharmacological agents have more than one target,” says Ehud Isacoff, a neurobiophysicist who holds joint appointments with Berkeley Lab’s Physical Biosciences Division and UC Berkeley’s Department of Molecular and Cell Biology. “Our study shows that the inhibition of TREK1 by fluoxetine, which was found in earlier studies, is accompanied by an unbinding of the protein’s C-terminal domain from the membrane. This is the first observation of the mechanism by which TREK1 might be regulated by antidepressant drugs.”

Guillaume Sandoz (seated) and Ehud Isacoff developed a fluorescent assay that enabled them to study TREK1 as a possible target for antidepressants. (Photo by Roy Kaltschmidt, Berkeley Lab Public Affairs)
Isacoff is the corresponding author on a paper reporting the results of this study that appears in the Proceedings of the National Academy of Science (PNAS). The paper is titled “Optical probing of a dynamic membrane interaction that regulates the TREK1 channel.” Co-authoring this paper were Guillaume Sandoz, a TREK1 specialist with France’s National Center for Scientific Research at the Institute for Molecular and Cellular Physiology, and PhD student Sarah Bell, both of whom were with Isacoff’s research group at the time the work was done.
Neurons in the human brain are like high-speed transistors, controlling the flow of electrical current through channels in their membranes by the opening and closing of molecular “gates” that control the flow of ions through selective pores. TREK1 is one of the most ubiquitous of these transmembrane proteins, gating the passage of potassium ions through neural membranes, which sets the excitability of the neuron. Earlier studies had shown that when the TREK1 gene is “knocked out” of mice, the mice display a depression-resistant phenotype that mimics the behavior of mice treated with fluoxetine and that the antidepressant inhibits the activity of the TREK1 channel. While these results pointed to a possible role for the TREK1 ion channel in the beneficial response to fluoxetine, the mechanism behind this activity was unclear.
“Studying what the different protein parts of an ion channel do is a huge challenge,” Isacoff says. “Over the years, my group has developed techniques by which the domains of channel proteins can be labeled with site-specific fluorescent dyes. Structural rearrangements of the labeled sites in the channel can then be detected through changes in the fluorescence.”
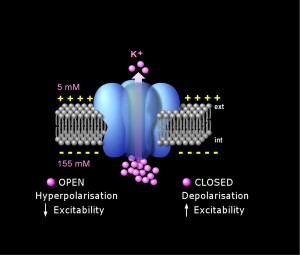
The TREK1 ion channel (blue) controls the passage of potassium ions (pink) through the plasma membrane (grey) of neurons, which sets neuron excitability. (Image from Isacoff group)
Isacoff and his group separated the C-terminal domain from the rest of the protein and tagged it with a green fluorescent protein (GFP) – a fluorescent protein from jellyfish commonly used to paint cells green for biological studies. Whereas the pore of the TREK1 ion channel is embedded in the plasma membrane of a neuron, the C-terminal is a short tail that protrudes out into the surrounding cytoplasm.
Using voltage clamps to measure electrical currents through the channel and fluorescence to monitor the disposition of the C-terminal domain, Isacoff and his group found that when the C-terminal tail is fully bound to the plasma membrane, the TREK1 potassium channel opens more; when the tail is unbound from the plasma membrane, the ion channel tends to close.
“We found that fluoxetine causes the isolated C-terminal domain to unbind from the membrane and also causes an inhibition of current from the full TREK1 channel,” Isacoff says.
The next step will be to see how the C-terminal tail is affected by the presence of fluoxetine when the tail is still attached to the rest of the TREK1 protein. In the meantime, Isacoff and his team feel they now have a valuable assay that can be used to monitor the reversible plasma membrane association of protein domains without the need for scanning, optical slicing or imaging.
“Pharmaceutical companies screening for potential new drugs, such as improved antidepressants, prefer assays that are fast and simple,” Isacoff says. “Our technique can be used to follow changes in lipid composition that result from membrane signaling events, or to study the binding to membranes by cytoplasmic regulatory domains of ion channels. This could be very useful for pharmaceutical research.”
This research was primarily supported by the National Institutes of Health.
Lawrence Berkeley National Laboratory is a U.S. Department of Energy (DOE) national laboratory managed by the University of California for the DOE Office of Science. Berkeley Lab provides solutions to the world’s most urgent scientific challenges including sustainable energy, climate change, human health, and a better understanding of matter and force in the universe. It is a world leader in improving our lives through team science, advanced computing, and innovative technology. Visit our Website at www.lbl.gov
Additional Information
The PNAS paper “Optical probing of a dynamic membrane interaction that regulates the TREK1 channel” can be viewed here: http://www.pnas.org/content/early/2011/01/18/1015788108.full.pdf
For more about the research of Ehud Isakoff, visit his Website at http://mcb.berkeley.edu/labs/isacoff/