Media contact: Paul Preuss, (510) 486-6249, [email protected]
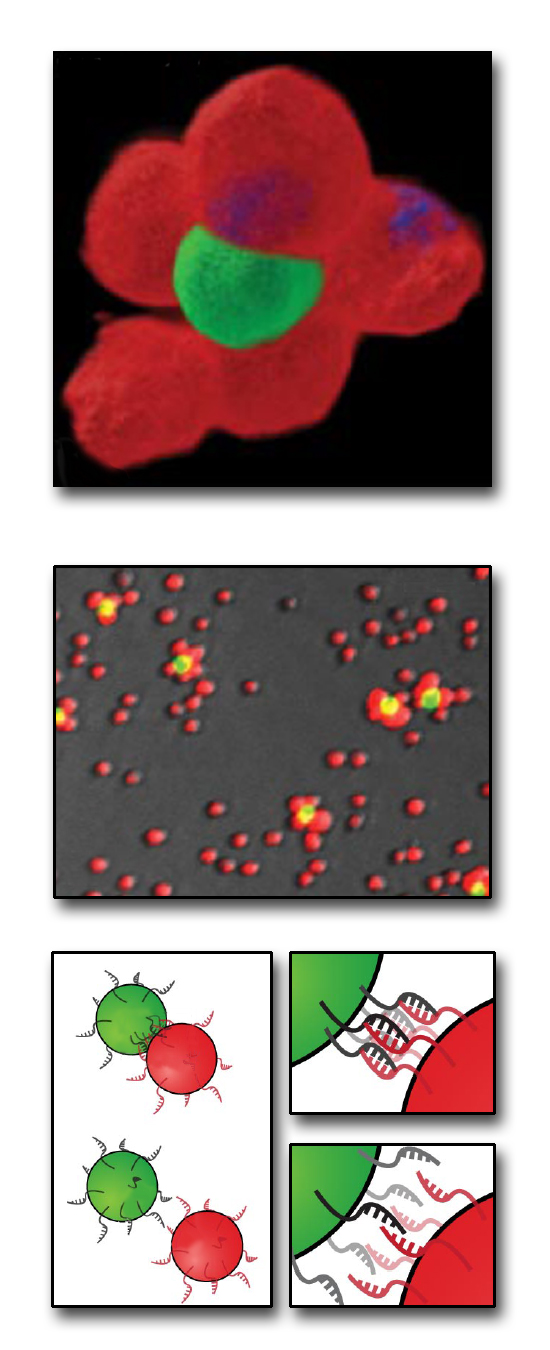
From the bottom up: how to build a microtissue. At bottom, cells bearing complementary single strands of DNA on their surfaces react with each other to form stable cell–cell contacts. At center are Jurkat cells stained red or green, labeled with different, complementary DNA sequences, and combined at a ratio of 50 to 1. At top, the two cell types are shown joined in a 3-D multicellular structure. (Click on image for best resolution.)
BERKELEY, CA – Scientists at the U.S. Department of Energy’s Lawrence Berkeley National Laboratory can now control how cells connect with one another in vitro and assemble themselves into three-dimensional, multicellular microtissues. The researchers demonstrated their method by constructing a tailor-made artificial cell-signaling system, analogous to natural cell systems that communicate via growth factors.
Artificial tissues are presently used in medicine for a range of applications such as skin grafts, bone marrow transplants, or blood substitutes, as well as in basic medical and biological research. Tissue engineers try to improve upon or repair natural tissues by manipulating living cells from one or more donors, sometimes in combination with synthetic materials. Unfortunately, in this “top down” approach, the cells assemble themselves randomly, losing the 3‑D organization that is key to many tissue functions.
“Our method allows the assembly of multicellular structures from the ‘bottom up,’” says Carolyn Bertozzi, principal investigator in the research, who directs DOE’s Molecular Foundry nanoscience research facility at Berkeley Lab and is a member of the Lab’s Materials Sciences and Physical Biosciences Divisions. “In other words, we can control the neighbors of each individual cell in a mixed population. By this method, it may be possible to assemble tissues with more sophisticated properties.”
An example of a state that depends on sophisticated cellular connectivity and communication is the stem-cell niche, “which can maintain stem cells in an undifferentiated state, or in some cases guide their differentiation down a specific pathway,” says Bertozzi, who is also a professor in the Departments of Chemistry and Molecular and Cell Biology at the University of California at Berkeley and a Howard Hughes Medical Institute investigator.
Bertozzi and her colleague Zev Gartner, who is now an assistant professor of Pharmaceutical Chemistry at the University of California at San Francisco, based their method of assembling 3‑D microtissues on an approach familiar to synthetic organic chemists, who build up novel molecular structures by combining one atom or functional group at a time. In the same way, Bertozzi and Gartner use synthetic methods to build microtissues one cell type at a time. Their results appear in the Proceedings of the National Academy of Sciences, PNAS Early Edition week of March 9, 2009.
Controlling cellular connections
The researchers enabled cells to react with other cells in a controlled way by coating the cell surfaces with DNA (not where DNA is normally found!). First they induced the cells to express artificial sugars bearing special chemical groups. Lengths of synthetic DNA, introduced into the cell-growth medium, were equipped to recognize these synthetic sugars on the cell surfaces and chemically bind to them.
The researchers coated cell surfaces from one group with strands of single-stranded DNA only 20 bases long, and the surfaces of another group with the complementary DNA strand. When a cell from one group meets its counterpart, the single strands recognize each other and form double-stranded DNA, which binds the cells together.
Bertozzi and Gartner discovered three variables that determine how cells from different groups react with one another. One is the ratio of the two kinds of cells: if both cell populations are equal, every cell finds a single partner and no complex assemblies form. But if there are, say, 50 times more cells from one group than from the other, numerous cells from the larger group will cluster around each cell from the smaller group.
Another variable is the complexity of the synthetic DNA sequence. The researchers can specify the complexity – for example, from a simple repeat of two bases such as cytosine and adenine (CACACA…) on one strand, which binds to a complementary repeating sequence of thymine and guanine (TGTGTG…), up to bases whose order varies over the full length of the 20-base strand of synthetic DNA. The more complex the sequences they display, the longer it takes the cells to bind together.
A third variable is the density of the DNA on the cell surface. By controlling how many artificial sugars the cells express, the researchers can control the DNA surface density. The greater the density, the faster the cells bind to one another.
Bertozzi says more variables for controlling cell assemblies are possible. “For example, it might be possible to cluster DNA strands on specific cellular structures. Thus, distribution of DNA on the cell surface might be yet another parameter we can exploit to guide cell-cell interactions.”
Building complex microtissues one step at a time
By controlling these variables to assemble small cellular structures, then separating the desired structures from unwanted ones and unreacted cells and assembling more cells on the purified collection – then repeating the steps again – the researchers can synthesize large, complex microtissues in much the same way a synthetic organic chemist assembles a complex molecule.
Bertozzi and Gartner applied these methods to build a signaling network where one kind of cell controls the growth of a second kind of cell. They maintained the survival and replication of hematopoietic progenitor cells (a kind of stem cell for blood cells), which depend on the presence of the growth factor interleukin-3, by combining them in microtissues with CHO cells (Chinese hamster ovary cells) that were engineered to secrete interleukin-3.
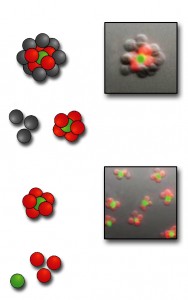
From the bottom up: after cell types labeled with red and green dye markers are joined, the resulting 3-D structures are purified to eliminate unreacted cells. More cells can then be added to form even more complex structures. There is no theoretical limit to the number of different cell types that can be assembled; microtissues with three or four different kinds of cells should be feasible. (Click on image for best resolution.)
When the two cell types were randomly mixed, the stem cells didn’t grow. But structured microtissues built from the two cell types stimulated their own growth, forming a simple artificial signaling network that behaved much like the natural networks that control immune-cell expansion or tumor proliferation.
“Since DNA has essentially an unlimited capacity for information storage, there is no theoretical limit on the number of different cell types we can assemble in a structure,” says Bertozzi. The key is to give each cell type its own unique DNA “bar code,” enabling its programmed interaction with any other specified cell type. “In practice, I think structures with three or four cell types are quite feasible. Such structures would be relevant to many biological organs.”
Structured microtissues have numerous research applications, says Bertozzi, particularly “in probing how the local cellular environment affects the behavior of a particular cell. Also, we can study how systems of cells work together to produce complex organ functions. Examples include how T cells and B cells work together in the lymph nodes to mount an immune response against foreign antigens.”
Practical challenges remain, such as scaling up the production of tailored microtissues to quantities needed for biomedical applications. Beyond that, Bertozzi hopes to refine the present method of modifying cell-surface DNA.
“As it stands, the need for unnatural sugar biosynthesis limits the kinds of cells that we can use in microtissue construction,” she says. “There are other ways in which DNA can be conjugated to cells, independent of their sugar metabolic pathways, and we intend to explore those avenues.”
“Programmed assembly of 3-dimensional microtissues with defined cellular connectivity,” by Zev J. Gartner and Carolyn R. Bertozzi, appears in PNAS Early Edition, week of March 9, 2009, and is available to subscribers at http://www.pnas.org/content/early/2009/03/06/0900717106.abstract?etoc. Research was supported by the Office of Science, U.S. Department of Energy.
Berkeley Lab is a U.S. Department of Energy national laboratory located in Berkeley, California. It conducts unclassified scientific research and is managed by the University of California. Visit our website at http://www.lbl.gov.