Contact: Lynn Yarris, (510)486-5375, [email protected]
BERKELEY, CA – The molecular architecture of a protein complex that helps determine the fate of human cells has been imaged for the first time by researchers with the U.S. Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab). Known as a human RISC-loading complex, this structure consists of snippets of ribonucleic acid (RNA) that control whether genetic messages are silenced or expressed.
From these new images, the research team, led by biochemist Jennifer Doudna and biophysicist Eva Nogales, has been able to propose a model of how RISC and other so-called “small RNA molecules” are able to target specific messenger RNA molecules for gene silencing and/or destruction. Their results have been published in the journal Nature Structure and Molecular Biology in a paper entitled: “Structural insights into RNA Processing by the Human RISC-Loading Complex.” RISC stands for RNA-Induced Silencing Complex.
Doudna and Nogales both hold joint appointments with Berkeley Lab, the University of California (UC) Berkeley, and the Howard Hughes Medical Institute (HHMI). Co-authoring the paper with them were Hong-Wei Wang, Cameron Noland, Bunpote Siridechadilok, David Taylor, Enbo Ma and Karin Felderer.
“We now know how the three main components of the RISC machinery – the Dicer and Argonaute enzymes and the TRBP binding protein – are arranged, and how they interact with one another and are likely to interact as a complex with messenger RNA,” says Doudna, an authority on RNA molecular structures. “Our work should help others in the design of mutants to test the mechanisms of the RNA binding and processing used by the gene-silencing RNA machinery in humans.”
Says Nogales, an expert on electron microscopy and image analysis, “Because of the relatively small size of RISC and the added complications of its not being very stable and having highly mobile parts, imaging this complex was a challenge. We used negative-stain electron microscopy and sophisticated single particle analysis.”
Human versions of small RNA molecules consist of approximately 20 to 30 nucleotides – compared to the three billion nucleotides in a molecule of human DNA – and include small interfering RNAs (siRNA) and microRNAs (miRNA). Despite their tiny size, small RNA molecules are receiving a great deal of attention from the biomedical and biotechnology communities. While some small RNA molecules help carry out genetic instructions, siRNAs and miRNAs work to silence genes and prevent them from issuing instructions that would be harmful to the cell such as “turn cancerous.”
In humans, targeted gene silencing, also known as RNA interference, requires that messenger RNA, the RNA that carries DNA’s genetic coding instructions, be bound or loaded with the RISC protein group. RISC will then either cleave the messenger RNA or block its message from being translated. This process is used in a number of important activities, including viral defense, chromatin remodeling, genome rearrangement, developmental timing, brain morphogenesis and stem cell maintenance.
Doudna, Nogales and their colleagues have identified the central component of the RISC-loading complex as the Dicer enzyme and that the human version is “L-shaped.” The short branch of the L-shaped human Dicer was shown to interact with RNA-binding protein TRBP, and the long branch of the L interacts with an Argonaute2 protein, which in turn binds with siRNA molecules (for cleaving messenger RNAs) or miRNA molecules (for blocking translation).
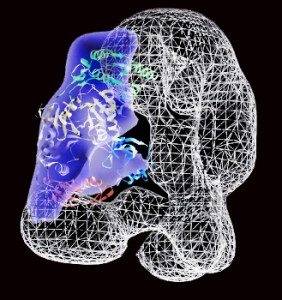
Electron microscopy structure of the human RISC-loading complex, with the L-shaped Dicer enzyme shown as a wire map and the Argonaute2 protein, shown in purple.
“Guide RNAs are generated from double-stranded RNA precursors by the enzyme Dicer, which binds directly to both Argonaute2 and TRBP,” says Doudna, whose earlier work showed how Dicer snips double-stranded RNA into segments of specific lengths. “Once loaded onto the Argonaute2 protein, a passenger strand is cleaved by Argonaute2 and dissociates from the complex, leaving behind a short single-stranded guide RNA that base-pairs with complementary messenger RNAs and targets them for catalyzed degradation or translational arrest by Argonaute2.”
Adds Nogales, “The results of this study indicate that the TRBP protein is flexibly linked to the Dicer enzyme, which has led to several specific hypotheses about the roles of these three proteins in binding and cleaving RNA during gene silencing by RNA interference.”
Doudna and her research group will be continuing this work with co-author by Hong-wei Wang, who carried out the microscopy and computation work for this study while he was a member of Nogales’ research group, and who is now an associate professor at Yale University.
“Hong-wei is investigating the dynamics of the proteins in the RISC-loading complex and the location of RNA in the complex at different steps in the RNA processing pathway,” says Doudna. “Together with higher-resolution structural studies, we should be able to provide even more new insights into the core machinery of RNA interference.”
This research was supported in part by grants from the U.S. National Institutes of Health and the Human Frontier Science Program.
Berkeley Lab is a U.S. Department of Energy national laboratory located in Berkeley, California. It conducts unclassified scientific research for DOE’s Office of Science and is managed by the University of California. Visit our Website at www.lbl.gov/
Additional Information
For more information about the Doudna research group see http://rna.berkeley.edu/
For more information about the Nogales research group see http://cryoem.berkeley.edu/
