Contact: Dan Krotz, [email protected], (510) 486-4019
Berkeley Lab scientists have developed a nanosized synthetic polymer bundle that can fold in half and trap a zinc molecule between its jaws, a first-of-its-kind feat that mimics how proteins conduct life’s vital functions.
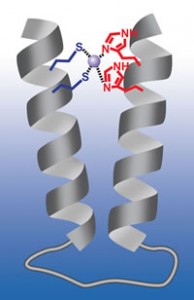
A more protein-like synthetic structure. Scientists have created a nano-sized polymer bundle that targets zinc, a development that opens the door for more rugged and longer lasting sensors and A more protein-like synthetic structure. Scientists have created a nano-sized polymer bundle that targets zinc, a development that opens the door for more rugged and longer lasting sensors and pharmaceuticals. In this schematic, a peptoid two-helix bundle clamps shut on a zinc molecule, in purple, locking the molecule in place.
The scientists’ success in coaxing protein-like function from a synthetic polymer is an initial step toward developing nanostructures that combine the precision of proteins with the ruggedness of non-natural materials. Although very primitive by nature’s standards, their polymer bundle could lead to highly accurate sensors capable of operating in harsh environments, or disease-targeting pharmaceuticals that last much longer than today’s therapies.
“We’re using nature as our guide to develop functional, stable nanostructures,” said Ron Zuckermann, who is the Facility Director of the Biological Nanostructures Facility in Berkeley Lab’s Molecular Foundry. Zuckermann developed the foldable polymer bundle with Byoung-Chul Lee and Tammy Chu of the Lab’s Materials Sciences Division, and Ken Dill of the Lab’s Physical Biosciences Division and the University of California at San Francisco, where he’s a professor of pharmaceutical chemistry.
“We have a long way to go, but the ultimate goal is to make useful nano-structured materials that can function over a wide range of conditions,” said Zuckermann.
It’s no surprise that Zuckermann and colleagues are working to emulate proteins. Millions of years of evolution have imbued them with unmatched molecular recognition and catalysis capabilities. Proteins have the ability to selectively bind with one — and only one — type of molecule. They also initiate incredibly precise chemical transformations, such as cutting a DNA strand in just the right place. If scientists can harness this laser-like targeting ability, they have the makings of an incredibly powerful way to attack disease and detect compounds.
But there’s a hitch. What proteins boast in precision, they lack in ruggedness and stability. They’re limited to narrow temperature and acidity ranges. They require a watery solution. And they degrade over time. These drawbacks limit their utility. Scientists already use proteins to target pathologies at the molecular scale, but the proteins degrade over time, curbing their effectiveness. Likewise, a protein-based sensor would be unsurpassed at sniffing out harmful contaminants, but it wouldn’t be able to operate in hot, cold, or dry conditions.
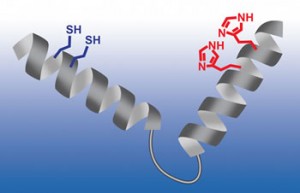
A set trap, waiting for zinc. Berkeley Lab scientists optimized the nano-sized structure to fold and bind with zinc by positioning zinc-binding residues, in red and blue, at just the right locations at each end.
“Our goal is to take proteins’ catalysis and molecular-recognition capabilities, and add them to a material that is more rugged and less prone to degradation,” said Zuckermann. “Proteins are precisely folded linear polymer chains of amino acids. So we thought, why not make a similar polymer chain by linking together non-natural amino acids?”
Specifically, his research team works with a protein-like chain of polymers called a peptoid. Peptoids are synthetic structures that mimic peptides, which nature uses to form complex proteins. Instead of using peptides to build proteins, however, Zuckermann’s team is striving to use peptoids to build synthetic structures that behave like proteins.
Their initial forays are promising. In previous work, they fashioned peptoids, one carefully placed monomer at a time, into one of nature’s most useful building blocks: a helical structure. They also linked two helix structures together using an unstructured segment in the middle. And they were able to fold this two-helix bundle in half, mimicking a change in shape that proteins use to conduct work.
From left, Ron Zuckermann, Tammy Chu and Byoung-Chul Lee in front of a peptoid synthesizer at the Molecular Foundry, which they used to develop nano-sized jaws that mimic proteins.
But the structure couldn’t do any of the tricks that proteins can, like target a molecule. To give it this function, the scientists set out to bind zinc. They chose zinc because the metal drives many fundamental biological processes, such as DNA recognition. If they could get the peptoid-based helical bundle to trap zinc, then they would have successfully mimicked a workhorse task performed by proteins.
To make this leap, they developed helical bundles with zinc-binding residues that are precisely positioned at both ends. They also added fluorescent tags at both ends, which allowed the scientists to measure when the bundles fold in half, trapping the zinc in place. The system worked: in some cases, the bundles captured zinc like a steel trap. The researchers varied the position and number of zinc-binding residues, tweaking the structure so it more readily snares zinc.
“It’s a team effort. Binding the zinc requires the contribution of both helices, and the zinc stabilizes the fold, firmly locking it in place,” said Zuckermann. “This is a triumph of chemists trying to mimic nature. It’s a first step toward being able to add biological functions, such as molecular recognition and catalysis, to a non-natural folded polymeric material.”
In the future, Zuckermann and colleagues hope to build more complex structures using three helices, which would yield a much more stable fold than two helices. The scientists’ research is detailed in a study entitled “Biomimetic Nanostructures: Creating a High-Affinity Zinc-Binding Site in a Folded Nonbiological Polymer” which was reported in the July 9, 2008 issue of the Journal of the American Chemical Society. The research was supported in part by the Department of Energy.