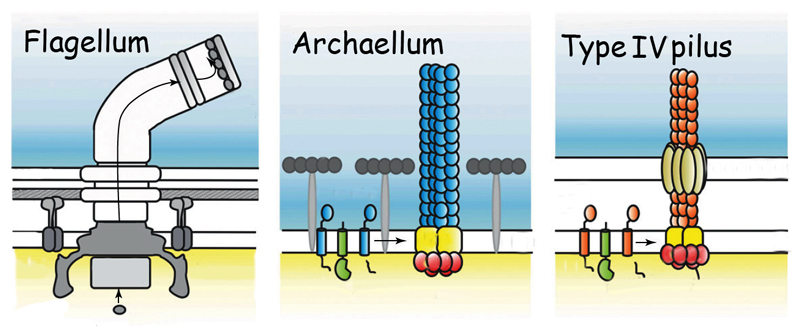
The motile structures in Bacteria and Archaea: the archaellum (center) functions like a bacterial flagellum but its structure resembles a bacterial Type IV pilus.
The protein structure of the motor that propels archaea has been characterized for the first time by a team of scientists from the U.S. Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) and Germany’s Max Planck Institute (MPI) for Terrestrial Microbiology.
The motility structure of this third domain of life has long been called a flagellum, a whip-like filament that, like the well-studied bacterial flagellum, rotates like a propeller. But although the archaeal structure has a similar function, it is so profoundly different in structure, genetics, and evolution that the researchers argue it deserves its own name: archaellum.
This unique motor is highly conserved in all motile archaeal species. Its structure most resembles that of the bacterial Type IV pilus, the filamentary “grappling hook” by which bacteria attach to surfaces and pull themselves along – and which is responsible for pathogenicity in many bacteria, including deadly strains of E. coli.
Since archaea may also be important players in the microbiota of the human gut, knowing the archaellum’s structure will help scientists understand how archaea interact with human cells. The Berkeley Lab-MPI research team reports its findings in the journal Molecular Cell.
Finding the key protein
Sulfolobus acidocaldarius was the model organism used in the analysis, says the research team’s co-leader Sonja-Verena Albers, who heads the MPI’s Molecular Biology of Archaea research group, “because this is one of the few well established model systems in which genetics works well. We have the genetic tools to mutate and precisely modify the Sulfolobus genome. We can combine in vivo experiments with the atomic structure of our proteins to see the effect of modifications.”
A protein called FlaI (pronounced “flah-eye”) was a leading candidate for archaella assembly and rotation, but the team had to find proof. FlaI is an ATPase – an enzyme that releases energy from adenosine triphosphate, or ATP – and was known to be involved in the assembly and function of Type IV pili in bacteria and the secretion of proteins in many microorganisms. But FlaI’s role in archaella was uncertain.
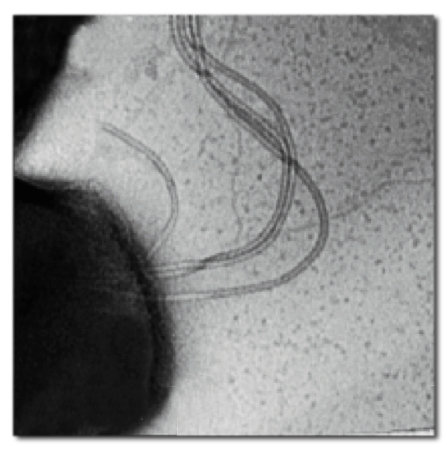
A wild-type S. acidocaldarius cell has only one to three archaella, so to see the results of deleting the FlaI gene the researchers created mutant strains with many archaella.
The Albers lab tested what happened to S. acidocaldarius when FlaI genes were deleted. Wild-type S. acidocaldarius cells have only one to three archaella on their surfaces, so “hyper-flagellated, hyper-motile” mutants with numerous archaella were created, to make deletion effects readily apparent. Deleting the FlaI gene eliminated the archaella and left the mutants unable to move. But when the researchers reintroduced the FlaI genes, the mutants were able to assemble archaella and use them to swim.
Now the challenge was to find out how FlaI performed its two functions, archaellum assembly and motility, by solving the FlaI protein structure. John Tainer of Berkeley Lab’s Life Sciences Division (LSD) and the Scripps Research Institute’s Department of Molecular Biology was co-leader of the research with Albers. The stable proteins of S. acidocaldarius, an extremophile able to withstand hot, acidic conditions, make it suitable for x-ray diffraction crystallography, and Tainer’s postdoctoral fellow, Sophia Reindl, was able to crystallize the FlaI protein.
To do the crystallography, Reindl used beamline 8.3.1 at Berkeley Lab’s Advanced Light Source (ALS), pinpointing the position of all the atoms in the FlaI protein and revealing that it consists of two parts. A globular C‑terminal domain, or CTD, is connected by a flexible linker to a more variable N‑terminal domain, or NTD, which constitutes a moveable tip.
As an energy-releasing enzyme the FlaI protein binds to an ATP nucleotide, from which it detaches a phosphate to generate energy – leaving adenosine diphosphate, ADP, in its place. After releasing the ADP, the protein binds to another ATP.
“Our goal was to solve as many structures as possible to see the protein in all its states and learn about its conformational changes,” Reindl says. “If you can find different orientations between the bound states of ATP and ADP, you can assume the protein is performing a certain movement – not that you can ever get thousand-percent proof, but you know it’s likely.”
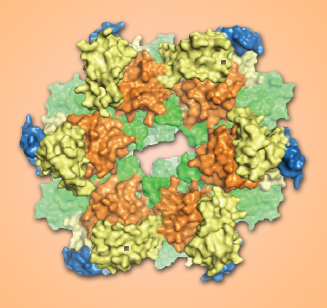
FlaI monomers arrange themselves into six-unit rings resembling a crown. Releasing a phosphate from ATP energizes a series of conformational changes that drive assembly and motility in the archaellum.
Reindl was able to crystallize FlaI bound to ADP, but not to ATP. Attempts to soak ATP into the crystals dissolved them, making x-ray crystallography of the ATP-bound state impossible.
“We then used SAXS at the ALS’s SIBYLS beamline to look at FlaI bound to ATP in solution,” she says. SAXS stands for small-angle x-ray scattering, and SIBYLS stands for Structurally Integrated Biology for Life Sciences, an ALS beamline maintained by the Life Sciences Division for which Tainer is director.
Says Reindl, “That had disadvantages – without a crystal we couldn’t get atomic resolution – but in some ways the advantages were greater, because we could see the overall conformation of the protein in solution, a more normal physiological state. By combining x-ray crystallography and SAXS data we could deduce how the structure changed.”
When bound to ATP, individual FlaI monomers arrange themselves into flat, six-unit rings, hexamers, with the ATPs serving as glue to hold them together. The result resembles a crown, with the CTD units forming the circlet and the free-to-move NTDs as the points.
Seven different conformations were recorded, revealing a dynamic play among the protein’s components in a changing, asymmetric assembly. From the detailed images, much of the action of the archaellum motor assembly could be deduced.
How FlaI builds an archaellum
The FlaI “crown” both assembles the archaellum and causes it to rotate, but it doesn’t work alone. Other important components are the protein FlaJ, which serves as a platform to which FlaI attaches and also forms a kind of bearing that penetrates the cell membrane, and FlaB, the subunits of the archaellum filament itself, plus other helper proteins.
“Just how ATPases catch the energy of ATP is a long-standing question,” says Tainer. The individual monomers in the FlaI crown process the ATP and reduce it to ADP by releasing a phosphate; here the “glue” that binds a FlaI monomer to its neighbor in the crown lets go. “In this phosphate release mechanism our team – including Abrahjyoti Ghosh and Gareth Williams in Life Sciences – were able to see a state never seen before, an intermediate conformation created by the released phosophate.”
Instantly the whole FlaI monomer in the crown moves upward, pushing up the FlaB filament and opening a gap at its base where waiting FlaB subunits are added to the filament, causing it to grow. The process is similar to the mechanism in a bacterial Type IV pilus, although the resulting structures operate very differently.
That FlaI is the protein uniquely responsible for both assembly and motility of the archaellum was established by further genetic studies. A mutant strain that lacked only the first 29 amino acids in the NTD point was quite capable of assembling archaella, but incapable of making them rotate.
“This mutant was interesting to us because it raised the question of how altered FlaI proteins keep the ability to assemble archaella, yet they lose their motility,” says Albers. “That the N-terminal-deletion mutant can assemble archaella, but the cells are not motile, implies that after assembly a switch is thrown, and the filament starts to rotate.”
The biggest challenge for the team’s continuing research is to learn how, as ATP releases energy, the upward movement of FlaI monomers is transferred to the rotation of the archaellum filament. Does the hexamer – the whole FlaI crown – rotate, or only the archaellum itself? And if so, how?
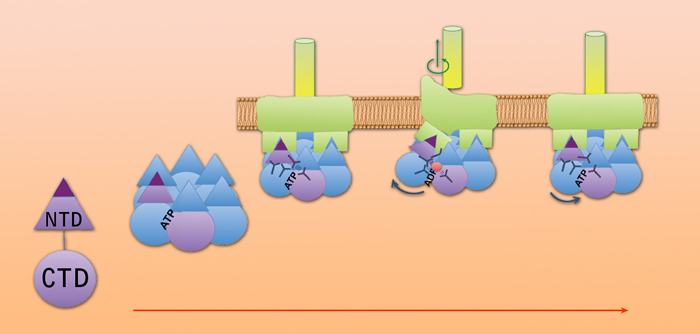
The story till now: individual FlaI proteins have two units, bases and points, connected by a flexible linker. When bound to ATP they assemble a six-member crown, which attaches to the base of the FlaJ protein in the cell membrane. When an individual FlaI protein hydrolyzes ATP and releases a phosphate, its point moves up and creates a space beneath the archaellum filament, where FlaB subunits can join the filament’s base, causing it to grow. The same changes in conformation drive rotation, but the exact mechanism is still be learned.
“A mechanistic understanding of microbial movements is critical to assessing their impacts on planet-scale recycling of elements, climate change, and medicine,” Tainer says. “We have studied Type IV pili systems for over a decade, including the ATPases needed for pathogen movement. Now this novel archaellum system allowed us to capture and genetically test a phosphate-release intermediate and other conformations that change our understanding of how the motors for all these broadly related microbial systems of movement and secretion work.”
While many questions remain, the MPI-Berkeley Lab collaboration has already produced the first and only in-depth structural study on an archaellum protein, another step toward solving the mysteries of Archaea, the third domain, besides Bacteria and Eukarya, of the Tree of Life.
The research was supported by National Institutes of Health grant AI22160, the Max Planck Institute, the Max Planck Society, and the Dutch Science Organization. The ALS is supported by DOE’s Office of Science, which also supports the SIBYLS beamline in partnership with NIH, grant GM105404.
###
“Insights on FlaI functions in archaeal motor assembly and motility from structures, conformations and genetics,” by Sophia Reindl, Abhrajyoti Ghosh, Gareth J. Williams, Kerstin Lassak, Tomasz Neiner, Anna-Lena Henche, Sonja-Verena Albers, and John A. Tainer, appears in Molecular Cell, online at http://www.cell.com/molecular-cell/abstract/S1097-2765%2813%2900047-6.
Visit Sonja-Verena’s laboratory at the Max Planck Institute for Terrestrial Biology at http://www.staff.uni-marburg.de/~albers/.
The SIBYLS beamline at the Advanced Light Source is at https://commons.lbl.gov/display/bl1231/The+SIBYLS+Beamline.
More information about how Archaea get around is at http://www.scoop.it/t/archaeal-surface-structures.
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
The Advanced Light Source is a third-generation synchrotron light source producing light in the x-ray region of the spectrum that is a billion times brighter than the sun. A DOE national user facility, the ALS attracts scientists from around the world and supports its users in doing outstanding science in a safe environment. For more information visit www-als.lbl.gov/.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit the Office of Science website at science.energy.gov/.