
Reginal Lamendella collecting samples. She was part of a team of Berkeley Lab scientists that analyzed how beach microbes reacted to the oil that washed ashore.
In June, 2010, two months after the Deepwater Horizon oil spill, Regina Lamendella collected samples along a hard-hit beach near Grand Isle, Louisiana. She was part of a team of Berkeley Lab researchers that wanted to know how the microbes along the shoreline were responding to the spill.
Some microorganisms love to consume hydrocarbons, and they’re known to thrive in the Gulf of Mexico. But were oil-hungry microbes also active on the beach?
The latest results are now in. As reported in a recent issue of the journal Frontiers in Microbiology, the scientists found a bloom in the abundance and diversity of microorganisms that have the ability to degrade oil.
Their research sheds light on a lesser-studied aspect of the environmental disaster. While numerous studies have explored what happened to the oil in the Gulf of Mexico, less is known about the fate of the oil that washed ashore early in the spill history.
Their work could also help scientists develop a “microbial fingerprint” of oil contamination for similar coastal ecosystems.
“We found that when the oil reached shore, it caused shifts in the microbial communities toward a hydrocarbon-degrading consortium,” says Lamendella, who conducted much of the work while a post-doctoral researcher in Berkeley Lab’s Earth Sciences Division. She is now a guest scientist at Berkeley Lab and an assistant professor of biology at Pennsylvania’s Juniata College.
Janet Jansson, a senior staff scientist in the Earth Sciences Division, led the research and is the corresponding author of the study.
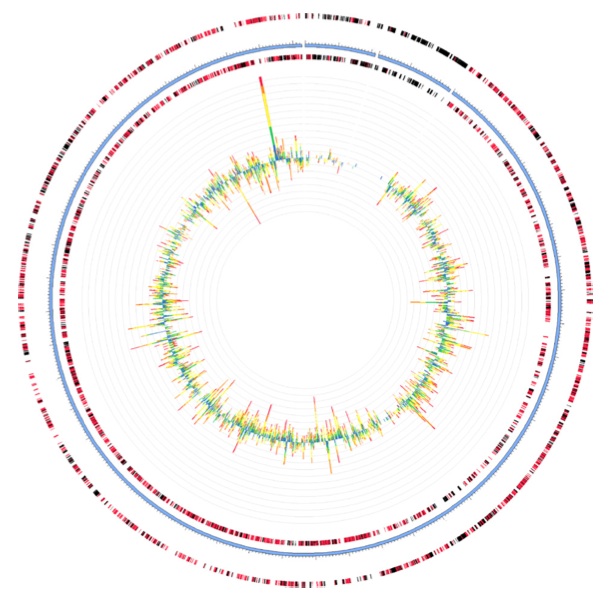
Here’s what a microbial response to oil looks like. The inner circle depicts the metatranscriptome of a sample collected on June 3, 2010. The outer circles depict the extent to which these expressed genes map to a microbe called Marinobacter aquaeolei strain VT8, one of the more prevalent species in the samples.
The scientists collected more than 150 oil-contaminated and uncontaminated samples in June 2010, while the oil accumulated on the beach. They then brought the samples to Berkeley Lab for analysis.
To find out what kinds of microbes were flourishing along the beach, they sequenced the 16S ribosomal RNA genes in the samples. These protein-making genes are found in all microbes, and in general each species has a unique variation. The scientists found that oil-contaminated samples had an abundance of 16S ribosomal RNA genes that corresponded to microbial families with members known to degrade hydrocarbons.
They also mapped the metatranscriptome present in the contaminated samples. A metatranscriptome is the “readout” of all of the genes active in a community at a given time. The data revealed the expression of several genes that degrade chemicals found in hydrocarbons. These genes are closely related to genes from oil-degrading species such as Alteromonadales, Rhodobacterales, and Pseudomonales.
They also isolated one of the most prevalent bacteria in the samples, and incubated it in the presence of oil obtained from the ruptured well. The experiments found that the bacterium, called Marinobacter, has the potential to transform hydrocarbons.
“We found a succession in the composition of microbial communities at the beach,” says Jansson. “This research may assist in the understanding of microbial proxies for oil contamination in similar coastal areas.”
Other Berkeley Lab scientists involved in the research include Sharon Borglin, Romy Chakraborty, Emmanuel Prestat, Neslihan Tas, and guest scientist Terry Hazen. Steven Strutt, an undergraduate student at Juniata College, conducted extensive bioinformatics work.
The research was funded by the Energy Biosciences Institute.
