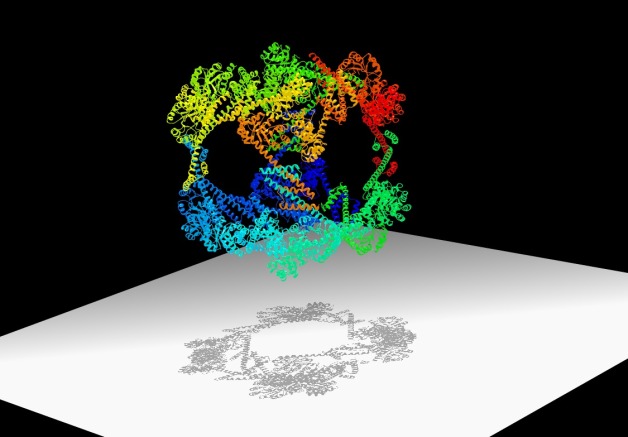
Scientists have designed a hollow, cube-shaped protein cage that has the potential to be a useful material for nanomedicine and other applications. Its crystal structure, shown here, was optimized at the Advanced Light Source.
With help from Berkeley Lab’s Advanced Light Source, scientists from UCLA recently designed a cage made of proteins.
The nano-sized cage could lead to new biomaterials and new ways to deliver drugs inside cells. It boasts a record breaking 225-angstrom outside diameter, the largest to date for a designed protein assembly. It also has a 130-angstrom-diameter central cavity, which is large enough to hold molecular cargo. And its high porosity is perfect for packing a lot of chemistry in a small package.
More research is needed, but perhaps scientists could some day insert a cancer-fighting drug inside the cage, and tweak its exterior proteins so that it targets malignant cells.
That’s one promise of the new cage. Another is the way in which it was designed. The cage is composed of specially designed “building block” proteins. When the proteins are in a solution with just the right conditions, they assemble into a hollow cube made of 24 proteins. Some of these cubes form crystals.
The scientists used the Advanced Light Source, a synchrotron located at Berkeley Lab, to quickly visualize the cage in different solutions. This helped the scientists determine how to best get the cage to assemble itself. It also allowed them to see how different solutions yield cages of various geometries.
They used beamline 12.3.1, also known as SIBYLS, which stands for Structurally Integrated Biology for Life Sciences. The SIBYLS beamline is optimized for the joint application of crystallography and SAXS imaging, or small-angle X-ray scattering. SAXS provides information on the shapes of large molecular assemblies in almost any type of solution. And it’s much faster than conventional protein crystallography techniques.
“SAXS helped us efficiently and quickly understand the assembly processes of these protein cages. We had feedback in a matter of hours, not days” says Greg Hura, a scientist with Berkeley Lab’s Physical Biosciences Division.
Hura and John Tainer of Berkeley Lab’s Life Sciences Division are co-authors of a Nature Chemistry paper that describes the protein cage. The research was led by Todd Yeates, a UCLA professor of chemistry and biochemistry.
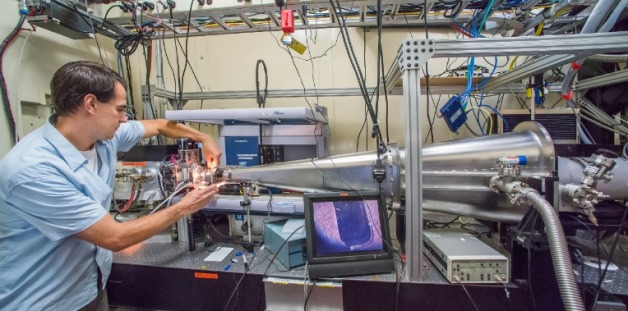
Greg Hura at the The SIBYLS beamline at the Advanced Light Source. The beamline can quickly visualize a protein assembly’s structure in almost any solution, and is helping researchers design new biomaterials.
SAXS made its mark elucidating the structure of proteins critical to human health, such as DNA repair machines. The technique can analyze about 100 samples in four hours. It also analyzes samples in solutions that approximate the biological conditions in which proteins are found. Hura and Tainer are now expanding SAXS’s repertoire to assist in the development of biomaterials.
“The magic of proteins is they are capable of a tremendous amount of chemistry, which we can harness in advanced materials for medicine, energy, and other applications,” says Hura, who helped optimize SAXS for high-throughput use.
The technique could be especially useful in helping to integrate the nanoscale properties of individual proteins into large complexes that perform useful functions. For example, Hura envisions using SAXS to develop protein assemblies that act as highly efficient catalysts, complete with millions of points that interact with a substance of choice.
“We are keenly interested in the rules for assembly at these nanoscales, since many alternative and valuable designs are currently being explored,” says Hura.
For the UCLA-developed protein cage, SAXS helped the scientists develop an annealing process that yielded crystal structures of the cage in eight hours. Before, it took several months for crystals to form. SAXS also enabled the team to analyze the protein cages under real-world physiological conditions, such as the pH levels found inside cells, and see how these conditions affected the cages’ properties.
“The technique allows the direct visualization of a structure’s flexibility and variability in solution, which will help improve the design of protein cages and other biomaterials,” says Hura.
