 A team of scientists at Berkeley Lab has developed an unsupervised multi-scale machine learning technique that can automatically and specifically capture biomedical events or concepts directly from raw data. In many data-driven biomedical studies, the data limitations (e.g., limited data scale, limited data label, unbalanced data and un-controllable experimental factors) impose great challenges to scientific discovery, which can only be addressed with advanced machine learning techniques. This work, described recently in IEEE Transactions on Pattern Analysis and Machine Intelligence, provides an effective and efficient way of learning and targeting sharable information so data can be used across domains. It also potentially removes limitations, especially for biomedical studies.
A team of scientists at Berkeley Lab has developed an unsupervised multi-scale machine learning technique that can automatically and specifically capture biomedical events or concepts directly from raw data. In many data-driven biomedical studies, the data limitations (e.g., limited data scale, limited data label, unbalanced data and un-controllable experimental factors) impose great challenges to scientific discovery, which can only be addressed with advanced machine learning techniques. This work, described recently in IEEE Transactions on Pattern Analysis and Machine Intelligence, provides an effective and efficient way of learning and targeting sharable information so data can be used across domains. It also potentially removes limitations, especially for biomedical studies.
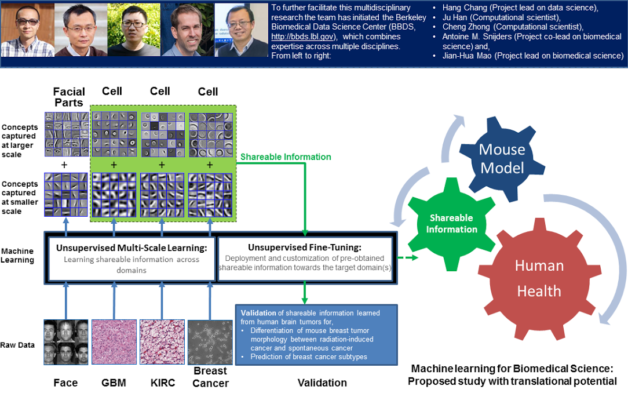
This multi-scale machine learning technique can be applied to many biomedical tasks, allowing the efficient and effective capturing of biomedical events or concepts at different scales (e.g., physical size) without any pre-defined biomedical endpoints or studies. An example of a pre-defined endpoint could be differentiation of tumor morphology that can predict metastatic risk. Researchers have shown that the information captured through this technique can be directly deployed or fine-tuned towards new endpoints or studies in related biomedical domains.
The core group in this team, Hang Chang, Antoine M. Snijders and Jian-Hua Mao, together with another scientist, Zhong Wang, of Berkeley Lab’s Biosciences Area, have initiated a Berkeley Biomedical Data Science Center (BBDS), which combines expertise across multiple disciplines to further facilitate and nurture data-intensive biomedical science. Hang Chang, a research scientist in the Lab’s Biological Systems & Engineering Division, said, “We have shown that our technique can be applied to other diverse biomedical tasks. For instance, the knowledge derived from human brain tumor histology can be directly utilized for the differentiation of mouse mammary tumor morphology between radiation-induced cancer and spontaneous cancer.” He added, “This suggests that our technique can be beneficial to biomedical studies with translational potential.”
The multi-scale machine learning technique helps improve the effectiveness and efficiency in learning sharable information across domains. Chang said, “When we determine basis information from data collected from cell culture or animal model studies, we think it will be possible to share and deploy the pre-attained information in human-related studies.” The BBDS plans to apply this technique to three ongoing projects related to cancer risk assessment of environment exposure, early stage cancer diagnosis and multi-modal biomarkers identification for personalized medicine. For more information, visit the BBDS website.