A protein used by common soil bacteria is providing new clues in the effort to convert aryl compounds, a common waste product from industrial and agricultural practices, into something of value.
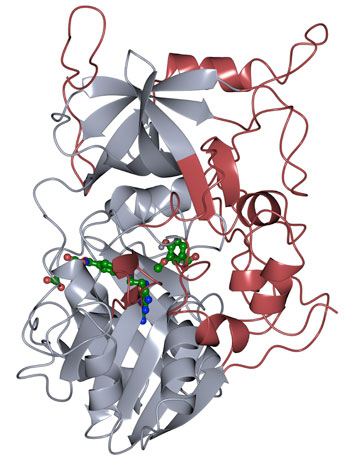
The protein structure of LigM was determined using X-ray crystallography, revealing novel structural elements that are unique to LigM (red) in addition to a conserved tetrahydrofolate-binding domain (gray) that is found throughout life. LigM binds to it’s substrates (green) using internal binding cavities. (Credit: Amanda Kohler/JBEI)
Researchers at the Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) and Sandia National Laboratories working at the Joint BioEnergy Institute (JBEI) have resolved the protein structure of the enzyme LigM, which is utilized by the soil bacterium Sphingomonas to metabolize aryl compounds derived from lignin, the stiff, organic material that gives plants their structure.
Their work is reported today in the Proceedings of the National Academy of Sciences.
In biofuel production, aryl compounds are a byproduct of the breakdown of lignin. Many of the pathways leading to the breakdown of lignin involve demethylation, which is often a critical precursor to any additional steps in modifying lignin-derived aryl compounds.
Study lead author Amanda Kohler, JBEI postdoctoral researcher at Sandia, noted that LigM is an attractive demethylase for use in aromatic conversion because it is a simple, single-enzyme system. LigM is also able to maintain its functionality over a broad temperature range.
“When we’re trying to build new pathways in synthetic biology, the simpler the system the better,” said Kohler.
The researchers found that half of the LigM enzyme was homologous to known structures with a tetrahydrofolate-binding domain that is found in simple and complex organisms alike. The other half of LigM’s structure is completely unique, providing a starting point for determining where its aryl substrate-binding site is located. They also figured out that LigM is a tyrosine-dependent demethylase.
“It’s the first of its kind to be identified,” said Kohler. “This research provides the much-needed groundwork to help in the development of an enzyme-based system for converting aromatic waste products into something useful.”

Postdoctoral researcher Amanda Kohler sets up enzyme reactions as part of JBEI’s Enzyme Optimization Group. (Credit: Marilyn Chung/Berkeley Lab)
Kohler said they are now working on engineering LigM so that it is able to act on a wider range of aryl substrates in addition to targeting specific aryl waste products.
Other JBEI study authors are Kenneth Sale and Matthew Mills from Sandia National Laboratories and Paul Adams and Blake Simmons from Berkeley Lab.
Funding from DOE’s Office of Science helped support this work. The researchers used Berkeley Lab’s Advanced Light Source, a DOE Office of Science User Facility, to collect diffraction data and to characterize the structure of LigM.
###
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel Prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.
