-Written By Lida Gifford
Cyclic proteins that assemble from multiple identical subunits (homo-oligomers) play key roles in many biological processes, including cell signaling and enzymatic catalysis and protein function. Researchers in Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging (MBIB) Division worked with University of Washington’s David Baker, who led a team to design in silico and crystallize self-assembling cyclic homo-oligomer proteins.
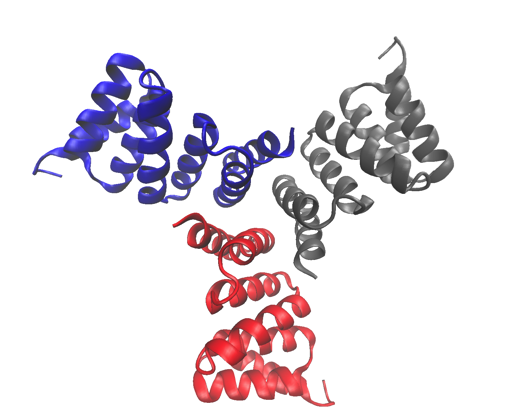
The high-resolution crystal structure of one of the proteins studied, a computationally designed trimer. The structure has been deposited in the Protein Data Bank with accession number 5HRZ.
By predicting and designing these higher-order oligomers, Baker and his team have gained an understanding of the fundamental principles underlying oligomer-oligomer interactions. Beyond the internal interactions of the protein itself, these designed oligomers can be used to explore basic questions about how the structure of signaling molecules affects the behavior of receptors and cellular response.
A strategy was developed to design interfaces onto idealized proteins aimed to direct their assembly into multimeric complexes. Berkeley Lab researchers used structural characterization—both X-ray crystallography and small angle X-ray scattering (SAXS)—to show that many of the designs adopted the target oligomerization state and predicted structure. Not only does their work demonstrate that scientists have a basic understanding of what determines oligomerization, it also shows that they can design proteins with tunable shape, size, and symmetry for a variety of biological applications.
Some of the X-ray crystallography work included in the paper was performed under the auspices of the Crystallography Collective program, which is run by research scientist Banumathi Sankaran at the Advanced Light Source. Fellow research scientist Henrique Pereira crystallized the proteins designed by the University of Washington researchers. Sankaran and Peter Zwart, MBIB staff scientist, collected crystallographic data on Beamline 5.0.2 in the Berkeley Center for Structural Biology and solved the structures. To gain information about the designed protein dynamics, Kathryn Burnett and Greg Hura of MBIB performed SAXS on the SIBYLS beamline.
Pereira, Sankaran, and Zwart have been co-authors on several papers with Baker and his team, all of which follow a theme of protein design and structure validation.
“Cyclic homo-oligomers play important roles in biological function,” said Sankaran. “Here we have another synthetic design that was proven to match the computational design with both small angle solution studies and X-ray scattering.”
This work, along with the curved beta sheet and trimeric metalloprotein designs, widen the possibilities for developing novel therapeutics and biomaterials.