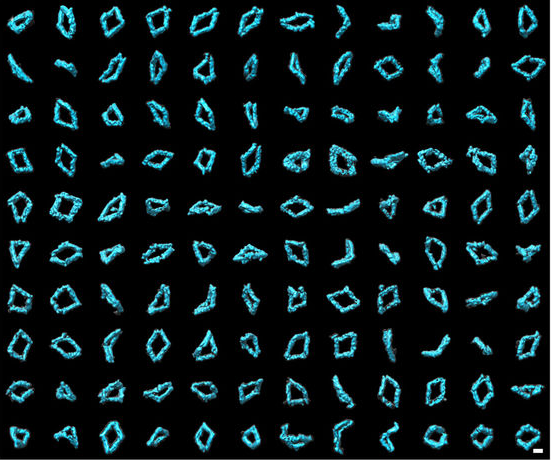
A collection of 120 3-D images of Bennett linkages. The images were created using individual-particle electron tomography (IPET), a technique developed at Berkeley Lab. (Credit: Lawrence Berkeley National Laboratory)
–By Laurie Chong
Over the past decade, researchers have been working to create nanoscale materials and devices using DNA as construction materials through a process called “DNA origami.”
A single long “scaffold” strand of DNA can be folded into specific shapes by hundreds of shorter strands, for example. The order of bases in each short strand acts as a blueprint that dictates the scaffold DNA’s final, 3-D shape.
Scientists hope to use this method to develop molecular devices that function as nanoscale machines or drug-delivery devices, markers for medical imaging or biological research, and components for electronic devices.
Long DNA scaffolds – composed of more than 10,000 DNA structural units called “nucleotides” – have remained difficult to manufacture and manipulate, which has so far limited the size of origami structures. It has also proven difficult to study the 3-D structure of these nanoparticles in their natural, flexible state.
Now, for the first time, a team of researchers from Lawrence Berkeley National Laboratory (Berkeley Lab) and Ohio State University have generated 3-D images from 129 individual molecules of flexible DNA origami particles. Their work provides the first experimental verification of the theoretical model of DNA origami.
The methods used here could easily be applied to other types of fabricated DNA origami structures, and help inform the design and optimization of future structures. Their work was recently published in Nature Communications.
The research team focused on DNA structures modeled after a basic mechanism called a “Bennett linkage,” which is a 3-D structure consisting of a chain of four rods connected by hinges. This creates a skewed quadrilateral shape in which the hinges are not parallel or in-line. Using Bennett linkages as building blocks, it’s possible to create expandable, useful structures, like supports for tents that can be rapidly assembled.

A schematic of a Bennett linkage and a model simulation showing the Bennett linkage made out of DNA. (Credit: Lawrence Berkeley National Laboratory)
DNA origami structures are very challenging to study because they are very flexible and delicate, and each molecule has a unique 3-D shape. This variability makes conventional imaging methods, like single-particle cryo-electron microscopy (cryo-EM), less suitable. Cryo-EM involves extensive computer “averaging” from thousands to hundreds of thousands of similar molecules. As a result, information about the flexible portions of DNA origami structures can be easily averaged away, yielding an incomplete picture of the structure.
The researchers relied on a technique developed at Berkeley Lab’s Molecular Foundry, a research facility for nanoscale science, to image the individual molecules that make up these structures. The method, called individual-particle electron tomography (IPET), takes pictures of a target molecule from multiple viewing angles, and then combines these pictures to create one 3-D, whole-molecule rendering, similar to how a medical computed tomography (CT) scan works.
Researchers captured 129 3-D images, with a resolution of 6 to 14 nanometers, that enabled them to tease out information about the dynamics and flexibility of DNA origami structures.
“The reconstructions confirmed that Bennett linkages have a high degree of structural diversity,” said Gang “Gary” Ren, staff scientist at the Molecular Foundry’s Imaging Facility and co-leader of the study.
A geometric analysis of these reconstructions shows that the conformations of Bennett linkage mechanisms are in good agreement with the theoretical models. When the linkages are near their “open” state, the “hinge” is almost fully extended. When the structures are nearer to their “closed” conformation, the structures take on different shapes and are extremely flexible and distorted.
“Based on these results and the model visualizations of the Bennett linkages, we can propose a new strategy for improving our control of Bennett linkages in large DNA scaffolds,” said Ren. “The approach includes redesigning the DNA sequences near the joints to stiffen the structure and prevent it from distorting near this joint.”
The Molecular Foundry is a DOE Office of Science User Facility.
In addition to Berkeley Lab scientists, other researchers contributing to this study were from Ohio State University. The work was supported by the National Science Foundation and the National Institutes of Health.
More information:
- Under the Electron Microscope – A 3-D Image of an Individual Protein
- Revealing the Fluctuations of Flexible DNA in 3-D
# # #
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel Prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.