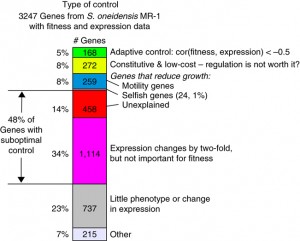
In a study of the microbe S. oneidensis MR-1’s ability to genetically adapt to environmental changes, Berkeley Lab researchers found that out of 3,247 protein-coding genes, only 5-percent showed a correlation between expression and fitness.
A new study challenges the orthodoxy of microbiology, which holds that in response to environmental changes, bacterial genes will boost production of needed proteins and decrease production of those that aren’t. Researchers with the U.S. Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab) found that for bacteria in the laboratory there was little evidence of adaptive genetic response. In fact, most bacterial genes appear to be regulated by signals unrelated to their function.
“Gene regulation in bacteria is usually described as an adaptive response to an environmental change so that genes are expressed only when they are required, but we’ve shown that in the laboratory gene regulation is often maladaptive,” says Adam Arkin, a systems and synthetic biologist and director of Berkeley Lab’s Physical Biosciences Division. “From our results, we propose that most bacterial genes are under indirect control, which means their expression is a response to signals not directly related to their function, and that their regulatory mechanisms perform poorly in the artificial conditions of a laboratory.”

Adam Arkin is a systems and synthetic biologist and director of Berkeley Lab’s Physical Biosciences Division. (Photo by Roy Kaltschmidt)
Arkin is the corresponding author along with Morgan Price, also with Berkeley Lab’s Physical Biosciences Division, of a paper describing this research in the journal Molecular Systems Biology. The paper is titled “Indirect and suboptimal control of gene expression is widespread in bacteria.” Other co-authors were Adam Deutschbauer, Jeffrey Skerker, Kelly Wetmore, Troy Ruths, Jordan Mar, Jennifer Kuehl and Wenjun Shao.
In the study of microbes, conventional scientific wisdom holds that bacterial genes are adaptively regulated to allow the microbe to respond to changing metabolic conditions or to environmental stresses. The idea is that expressing proteins provides a fitness benefit at a cost of cellular resources, therefore, under a cost-benefit model of optimal gene expression, proteins are up-regulated as required, and those that would add no benefit or be detrimental are down-regulated.
“Based on earlier work, we had this idea that bacterial regulation might not be as adaptive as most people seemed to think but we had no direct evidence,” says Price. “Once we had the technology, developed by Adam Deutschbauer, to measure fitness benefits on a large scale in bacteria, we were of course curious as to whether expression and fitness would be correlated.”
To test their ideas, Arkin, Price and Deutschbauer undertook an expression/fitness study of the MR-1 strain of Shewanella oneidensis, a bacterium that can reduce toxic heavy metals under both aerobic and anaerobic conditions.
“S. oneidensis MR-1 is important to DOE’s mission because of its potential applications in bioenergy and bioremediation,” says Deutschbauer. “Also, we were able to leverage an existing whole-genome mutant collection in S. oneidensis MR-1, which Adam Arkin’s research group had previously developed as part of a separate project.”
Adam Deutschbauer and Morgan Price were key contributors to a study that challenges the view that in response to environmental changes, bacterial genes boost production of needed proteins and decrease production of those that aren’t. (Photo by Roy Kaltschmidt)
The Berkeley Lab researchers collected genome-wide expression and fitness data for 3,247 of S. oneidensis MR-1’s protein-coding genes through several generations of the bacteria cultured under 15 different environments, including a variety of carbon sources, both aerobic and anaerobic conditions, and with and without the presence of toxic compounds. They also collected gene expression data for wild-type bacteria cultured under the same conditions. Overall, only five-percent of the examined S. oneidensis MR-1 genes showed adaptive regulation, whereas 48-percent showed suboptimal, non-adaptive regulation. Furthermore, 24-percent of the genes were highly expressed under conditions in which they were actually detrimental to fitness.
“If you simply plot how much a gene changes in expression in a given condition versus how much worse the cell grows without the gene in that condition over a large number of conditions, you find very little of the correlation you would expect under the traditional view of adaptive regulation,” says Arkin. “While we suspect adaptive regulation will line up better under more natural conditions, the fact that so few genes responded as expected in the laboratory indicates that whatever the natural response is, it probably will not fit the classical all-benefit- and-no-cost model.”
Similar expression/fitness analyses were performed on Zymomonas mobilis ZM4, an ethanol-producer, and Desulfovibrio alaskensis G20, a sulfate-reducer, with similar results to the S. oneidensis MR-1 study. The findings suggest that non-adaptive regulation of bacterial genes is widespread at least in laboratory settings.
“Bacteria have far more operons than sensors so they’re probably not well-designed to provide the feedback that the optimal regulation of most genes would require,” Price says. “Also, bacterial gene regulatory systems have evolved under very different conditions than those being tested in the laboratory. If the utility of a gene’s activity correlates with a functionally unrelated signal, then regulation by that signal will be selected for in the wild but probably not be maintained in artificial conditions. In fact, when we put the bacteria into an artificial situation, the regulatory system often becomes downright maladaptive.”
Price also believes that for maybe 10-percent of bacterial genes, there is little selective pressure to regulate their activity.
“These genes are always on at a constant low level so they are always beneficial or at least not very harmful,” he says. “There’s not much to gain from regulating their activity.”
Arkin, Price and Deutschbauer plan to extend their studies to approximately 100 additional types of bacterium. They also plan to carry out future studies under more naturalistic conditions.
“In addition to needing to test these systems under more natural conditions, there are a number of theories to explain the regulation of bacterial genes that we need to test,” says Arkin. “For example, it may be that transient gene expression changes are more indicative of response to environmental change than the long-term expression/fitness factors we measured, or there is more anticipatory control than we were able to test for in this study.”
This research was carried out through ENIGMA, a multi-institutional consortium funded by DOE through its Scientific Focus Area grant program. ENIGMA stands for Ecosystems and Networks Integrated with Genes and Molecular Assemblies. Its mission is to advance our understanding of microbial biology and the impact of microbial communities on their ecosystems.
# # #
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit the Office of Science website at science.energy.gov/.
Additional Information
For more about ENIGMA, go here
For more about the research of Adam Arkin, go here