The structure of a protein that is sending electrical pulses between neurons in your brain as you read this article has been fully mapped for the first time using Lawrence Berkeley National Laboratory’s Advanced Light Source. This much-anticipated milestone could lead to new treatments for neurological diseases and a better understanding of how the nervous system controls movement, memory, and learning.
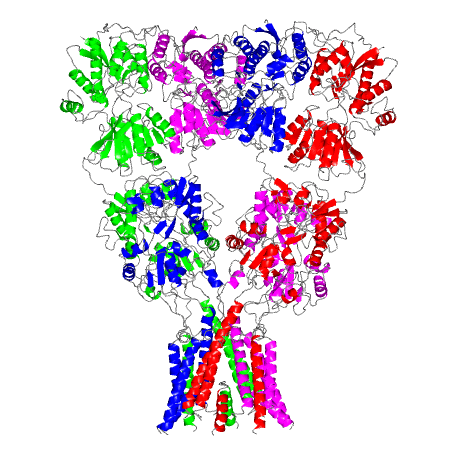
This computer-generated model of a rat glutamate receptor is the first complete portrait of this important link in the nervous system. At the top of the Y-shaped protein, a pair of molecules splay outward like diverging prongs. The bottom section forms the ion channel. The resolution of this image is 3.6 angstroms per pixel, or just under four ten-billionths of a meter per image unit.
The complete atomic-level architecture of the protein, called a glutamate receptor, caps more than 11 years of painstaking work by a team of scientists led by Eric Gouaux of the Oregon Health and Science University. The team’s research was featured on the cover of the December 10, 2009 issue of the journal Nature.
Glutamate receptors are lodged in the membranes of neurons. When a chemical neurotransmitter called glutamate attaches to the receptor, a channel opens in the membrane that allows ions to stream through. In this way, chemical signals are converted to electrical signals capable of being relayed between neurons and along nerves.
Although glutamate receptors and other neurotransmitter receptors are linchpins of the nervous system, their structure was poorly understood until now.
“This is the first blueprint of this incredibly important receptor. No one knew what it looked like,” says Gouaux, a pioneer in determining the atomic structure of neurotransmitter receptors and transporters.
With the three-dimensional structure of the protein now in hand, scientists can begin to explore how it mediates neural traffic: how does it help people learn new tasks, form fresh memories, and ride a bicycle? Scientists can also study what happens when the protein breaks down, a neural ‘pothole’ that is believed to contribute to Alzheimer’s, schizophrenia, and other neurodegenerative diseases.
“Knowing the receptor’s structure will help scientists design molecular therapies that fight disease by altering the receptor’s function,” explains Gouax. “It also gives everyone who studies the brain a model of how the many kinds of glutamate receptors are structured. It provides a new window into how the brain works.”
One of the scientists’ final stops on their journey to determine the protein’s structure was the Advanced Light Source, a national user facility located at Berkeley Lab that generates intense light for scientific research.

Keeping pace with genomics. Berkeley Lab's Peter Zwart at the Advanced Light Source's beamline 5.0.2, which is equipped to churn out atomic-scale resolution images of proteins.
The team started with a rat glutamate receptor, which was then crystallized and brought it to the Advanced Light Source, where it was exposed it to a beam of X-rays. The pattern in which X-rays scatter off of the atoms in the protein crystal was measured by a detector. This data was then translated by a computer program into a three-dimensional model of the protein.
This technique, called protein X-ray crystallography, was conducted at one of five Advanced Light Source beamlines operated by the Berkeley Center for Structural Biology.
“The glutamate receptor was characterized at beamline 5.0.2, which is our brightest beamline and is ideally suited to determine the structure of proteins at an atomic-level resolution,” says Peter Zwart of Berkeley Lab’s Physical Biosciences Division. “The number of photons in the beam is very high. This is important because more photons reaching the crystal means a stronger signal and a correspondingly shorter time to conduct the experiment.”
Such speed is needed to keep pace with the growing flood of proteins being generated by genomic studies of species and environmental samples. Each protein produced by a newly discovered gene must be structurally characterized in order to determine its function. To accommodate this demand, the Berkeley Center for Structural Biology boasts several tools that enable fast and efficient protein crystallography.
At beamline 5.0.2, for example, a robot automatically mounts and aligns protein crystals in the beamline. This enables faster data acquisition and eliminates the human errors that inevitably crop up in a repetitive procedure.
In addition, after the crystallographic data is obtained at the beamline, it is fed to a computer program called PHENIX that quickly generates a three-dimensional model of the protein. The program extracts the best data from the X-ray measurements, which it uses to piece together the protein’s atom-by-atom structure. It was developed by an international collaboration of scientists led by Paul Adams, the acting director of Berkeley Lab’s Physical Biosciences Division.
Take a 3-D spin of a glutamate receptor, which helps relay electrical signals between neurons and along nerves.
Additional information:
- Read the Nature paper “X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor.”
- More about the Advanced Light Source and the Berkeley Center for Structural Biology
- More about PHENIX
- More about Eric Gouaux and his research