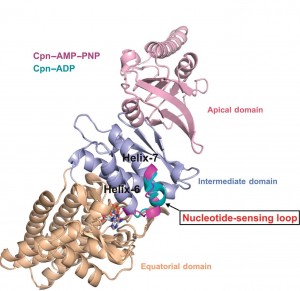
Berkeley Lab researchers at the Advanced Light Source have discovered a nucleotide-sensing loop that synchronizes conformational changes in the three domains of group II chaperonin for the proper folding of other proteins.
The gold standard for nanotechnology is nature’s own proteins. These biomolecular nanomachines – macromolecules forged from peptide chains of amino acids – are able to fold themselves into a dazzling multitude of shapes and forms that enable them to carry out an equally dazzling multitude of functions fundamental to life. As important as protein folding is to virtually all biological systems, the mechanisms behind this process have remained a mystery. The fog, however, is being lifted.
A team of researchers with the U.S. Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab), using the exceptionally bright and powerful x-ray beams of the Advanced Light Source, have determined the crystal structure of a critical control element within chaperonin, the protein complex responsible for the correct folding of other proteins. The incorrect or “misfolding” of proteins has been linked to many diseases, including Alzheimer’s, Parkinson’s and some forms of cancer.
“We identified, for the first time, a region within group II chaperonins we call the nucleotide-sensing loop, which detects the presence of the ATP molecules that fuel the chaperonin folding motion,” says Paul Adams, a bioengineer with Berkeley Lab’s Physical Biosciences Division and leading authority on x-ray crystallography who led this work. ““We knew that ATP hydrolysis is important for promoting protein folding, but we did not know how ATP activity was sensed and communicated.”
Adams is the corresponding author of a paper in The EMBO Journal that describes this study, which was performed in collaboration with colleagues at MIT and Stanford. The paper is titled “Mechanism of nucleotide sensing in group II Chaperonins.” Co-authoring this paper were Jose Pereira, Corie Ralston, Nicholai Douglas, Ramya Kumar, Tom Lopez, Ryan McAndrew, Kelly Knee, Jonathan King and Judith Frydman.

(From left) Paul Adams, Corie Ralston, Jose Henrique Pereira and Ryan McAndrew at the Advanced Light Source where they used the facilities of the Berkeley Center for Structural Biology to reveal important new information on protein folding. (Photo by Roy Kaltschmidt, Berkeley Lab)
Chaperonins promote the proper folding of newly translated proteins and proteins that have been stress-denatured – meaning they’ve lost their structure – by encapsulating them inside a protective chamber formed from two rings of molecular complexes stacked back-to-back. There are two classes of chaperonins, group I found in prokaryotes; and group II found in eukaryotes. Much of the basic architecture has been evolutionarily preserved across these two classes but they do differ in how the protective chamber is opened to accept proteins and closed to fold them. Whereas group I chaperonins require a detachable ring-shaped molecular lid to open and close the chamber, group II chaperonins have a built-in lid.
“We obtained crystal structures at sufficient resolution to allow us to examine, in detail, the effects that changes in nucleotides states have on ATP binding and hydrolysis in group II chaperonins,” Adams says. “From these structures we see that the nucleotide-sensing loop monitors ATP binding sites for changes and communicates this information throughout the chaperonin. Functional analysis further suggests that the nucleotide-sensing loop region uses this information to control the rate of ATP binding and hydrolysis, which in turn controls the timing of the protein folding reaction.”
The double-ring chaperonin complex features multiple subunits that are grouped into three domains – apical, intermediate and equatorial. For group II chaperonins, the closing of the lid for protein-folding causes all three domains to rotate as a single rigid body, resulting in conformational changes to the chamber that enable the proteins within to be folded. The synchronized rotation of the chaperonin domains is dependent upon the communication to all the subunits that is provided by the nucleotide-sensing loop. In identifying the nucleotide-sensing loop and its controlling role in group II chaperonin protein-folding, Adams and his colleagues may have opened a new avenue by which modified protein-folding activities could engineered.
“The strong relationship between incorrectly folded proteins and pathological states is well documented,” Adams says. “Since ATP hydrolysis is required for protein folding, it could be possible to engineer a nucleotide-sensing loop that promotes slower or faster protein folding activity in a given chaperonin. This could, for example, be used to increase the protein folding activity of human chaperonin, or perhaps reduce the cellular accumulation of misfolded proteins that can cause disease and other problems.”

Overlooking the San Francisco Bay, Berkeley Lab’s Advanced Light Source is a DOE Office of Science national user facility providing premier beams of x-ray and ultraviolet light for scientific research. (Photo by Roy Kaltschmidt, Berkeley Lab)
A key factor that enabled Adams and his colleagues to solve the three-dimensional crystal structure of the nucleotide sensing loop and determine its pivotal role in the protein folding of group II chaperonins was the unique protein crystallography capabilities of the Berkeley Center for Structural Biology. The BCSB operates five protein crystallography beamlines for Berkeley Lab’s Advanced Light Source (ALS), a DOE Office of Science national user facility for synchrotron radiation, and the first of the world’s third generation light sources. For this particular study, Adams and his colleagues used ALS beamlines 8.2.1 and 8.2.2, which are powered by a superconducting bending magnetic to yield higher energy x-rays that are optimized for the study of single crystals of biological molecules.
In this study, Adams and his colleagues studied an archaeon chaperonin. In their followup research, they will apply what they have learned to study the chaperonin known as TRiC, which is the human chaperonin.
“We believe that chaperonins have evolved to work on specific substrates and that the rates of protein folding may vary greatly between chaperonins in different organisms,” Adams says. “The structural and biochemical identification of the changes related to ATP hydrolysis provides important insights into the complex puzzle of protein folding for each type of chaperonin.”
This work was performed as part of the Center for Protein Folding Machinery, an NIH Roadmap-supported Nanomedicine Development Center. The BCSB is supported in part by the National Institutes of Health and National Institute of General Medical Sciences, and the Howard Hughes Medical Institute.
# # #
Lawrence Berkeley National Laboratory addresses the world’s most urgent scientific challenges by advancing sustainable energy, protecting human health, creating new materials, and revealing the origin and fate of the universe. Founded in 1931, Berkeley Lab’s scientific expertise has been recognized with 13 Nobel prizes. The University of California manages Berkeley Lab for the U.S. Department of Energy’s Office of Science. For more, visit www.lbl.gov.
Additional Information
For more information about Berkeley Lab’s Advanced Light Source go here
For more information about the Berkeley Center for Structural Biology go here
A YouTube video about the Berkeley Center for Structural Biology at the Advanced Light Source can be viewed here